| Bos taurus Gene: CDK6 | |||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Summary | |||||||||||||||||||||||||||||||||
| InnateDB Gene | IDBG-629049.3 | ||||||||||||||||||||||||||||||||
| Last Modified | 2014-10-13 [Report errors or provide feedback] | ||||||||||||||||||||||||||||||||
| Gene Symbol | CDK6 | ||||||||||||||||||||||||||||||||
| Gene Name | cyclin-dependent kinase 6 | ||||||||||||||||||||||||||||||||
| Synonyms | |||||||||||||||||||||||||||||||||
| Species | Bos taurus | ||||||||||||||||||||||||||||||||
| Ensembl Gene | ENSBTAG00000044023 | ||||||||||||||||||||||||||||||||
| Encoded Proteins |
cyclin-dependent kinase 6
|
||||||||||||||||||||||||||||||||
| Protein Structure | |||||||||||||||||||||||||||||||||
| Useful resources | Stemformatics EHFPI ImmGen | ||||||||||||||||||||||||||||||||
| InnateDB Annotation from Orthologs | |||||||||||||||||||||||||||||||||
| Summary |
[Homo sapiens] CDK6 expression is induced upon LPS stimulation via TLR4-signalling pathway. CDK6 mediates the increased adhesion of macrophages in response to LPS challenge, and is important for LPS lethality. (Demonstrated in murine model)
[Mus musculus] Cdk6 expression is induced upon LPS stimulation via Tlr4-signalling pathway. Cdk6 mediates the increased adhesion of macrophages in response to LPS challenge, and is important for LPS lethality.
|
||||||||||||||||||||||||||||||||
| Entrez Gene | |||||||||||||||||||||||||||||||||
| Summary |
This gene does not have any Entrez summary - the following is the summary from its human ortholog ENSG00000105810:
The protein encoded by this gene is a member of the cyclin-dependent protein kinase (CDK) family. CDK family members are highly similar to the gene products of Saccharomyces cerevisiae cdc28, and Schizosaccharomyces pombe cdc2, and are known to be important regulators of cell cycle progression. This kinase is a catalytic subunit of the protein kinase complex that is important for cell cycle G1 phase progression and G1/S transition. The activity of this kinase first appears in mid-G1 phase, which is controlled by the regulatory subunits including D-type cyclins and members of INK4 family of CDK inhibitors. This kinase, as well as CDK4, has been shown to phosphorylate, and thus regulate the activity of, tumor suppressor protein Rb. Expression of this gene is up-regulated in some types of cancer. Multiple alternatively spliced variants, encoding the same protein, have been identified. [provided by RefSeq, Nov 2009] |
||||||||||||||||||||||||||||||||
| Gene Information | |||||||||||||||||||||||||||||||||
| Type | Protein coding | ||||||||||||||||||||||||||||||||
| Genomic Location | Chromosome 4:9792303-10039520 | ||||||||||||||||||||||||||||||||
| Strand | Reverse strand | ||||||||||||||||||||||||||||||||
| Band | |||||||||||||||||||||||||||||||||
| Transcripts |
|
||||||||||||||||||||||||||||||||
| Interactions | |||||||||||||||||||||||||||||||||
| Number of Interactions |
This gene and/or its encoded proteins are associated with 0 experimentally validated interaction(s) in this database.
They are also associated with 96 interaction(s) predicted by orthology.
|
||||||||||||||||||||||||||||||||
| Gene Ontology | |||||||||||||||||||||||||||||||||
Molecular Function |
|
||||||||||||||||||||||||||||||||
| Biological Process |
|
||||||||||||||||||||||||||||||||
| Cellular Component |
|
||||||||||||||||||||||||||||||||
| Orthologs | |||||||||||||||||||||||||||||||||
|
Species
Homo sapiens
Mus musculus
|
Gene ID
Gene Order
|
||||||||||||||||||||||||||||||||
| Pathway Predictions based on Human Orthology Data | |||||||||||||||||||||||||||||||||
| NETPATH |
TGF_beta_Receptor pathway
Wnt pathway
BCR pathway
|
||||||||||||||||||||||||||||||||
| REACTOME |
Cyclin D associated events in G1 pathway
G1 Phase pathway
Cellular responses to stress pathway
Oncogene Induced Senescence pathway
Mitotic G1-G1/S phases pathway
Cell Cycle pathway
Cellular Senescence pathway
Cell Cycle, Mitotic pathway
Senescence-Associated Secretory Phenotype (SASP) pathway
Oxidative Stress Induced Senescence pathway
Oxidative Stress Induced Senescence pathway
Cell Cycle, Mitotic pathway
G1 Phase pathway
Cellular responses to stress pathway
Oncogene Induced Senescence pathway
Cell Cycle pathway
Cyclin D associated events in G1 pathway
Cellular Senescence pathway
Senescence-Associated Secretory Phenotype (SASP) pathway
Mitotic G1-G1/S phases pathway
|
||||||||||||||||||||||||||||||||
| KEGG |
Non-small cell lung cancer pathway
Cell cycle pathway
Melanoma pathway
Glioma pathway
Small cell lung cancer pathway
Chronic myeloid leukemia pathway
Pancreatic cancer pathway
p53 signaling pathway pathway
Pathways in cancer pathway
p53 signaling pathway pathway
Melanoma pathway
Cell cycle pathway
Glioma pathway
Small cell lung cancer pathway
Chronic myeloid leukemia pathway
Non-small cell lung cancer pathway
Pancreatic cancer pathway
Pathways in cancer pathway
|
||||||||||||||||||||||||||||||||
| INOH |
IL-7 signaling pathway
JAK STAT pathway and regulation pathway
EPO signaling pathway pathway
VEGF signaling pathway pathway
IL-7 signaling pathway
JAK STAT pathway and regulation pathway
EPO signaling pathway pathway
VEGF signaling pathway pathway
|
||||||||||||||||||||||||||||||||
| PID NCI |
Coregulation of Androgen receptor activity
p73 transcription factor network
C-MYB transcription factor network
IL2 signaling events mediated by STAT5
|
||||||||||||||||||||||||||||||||
| Cross-References | |||||||||||||||||||||||||||||||||
| SwissProt | |||||||||||||||||||||||||||||||||
| TrEMBL | E1BC36 | ||||||||||||||||||||||||||||||||
| UniProt Splice Variant | |||||||||||||||||||||||||||||||||
| Entrez Gene | 511754 | ||||||||||||||||||||||||||||||||
| UniGene | Bt.25462 | ||||||||||||||||||||||||||||||||
| RefSeq | NM_001192301 XM_005205135 XM_005205136 XM_005205137 XM_005205138 | ||||||||||||||||||||||||||||||||
| HUGO | HGNC:1777 | ||||||||||||||||||||||||||||||||
| OMIM | |||||||||||||||||||||||||||||||||
| CCDS | |||||||||||||||||||||||||||||||||
| HPRD | |||||||||||||||||||||||||||||||||
| IMGT | |||||||||||||||||||||||||||||||||
| EMBL | DAAA02009765 | ||||||||||||||||||||||||||||||||
| GenPept | |||||||||||||||||||||||||||||||||
| RNA Seq Atlas | 511754 | ||||||||||||||||||||||||||||||||
| Transcript Frequencies | |||||||||||||||||||||||||||||||||
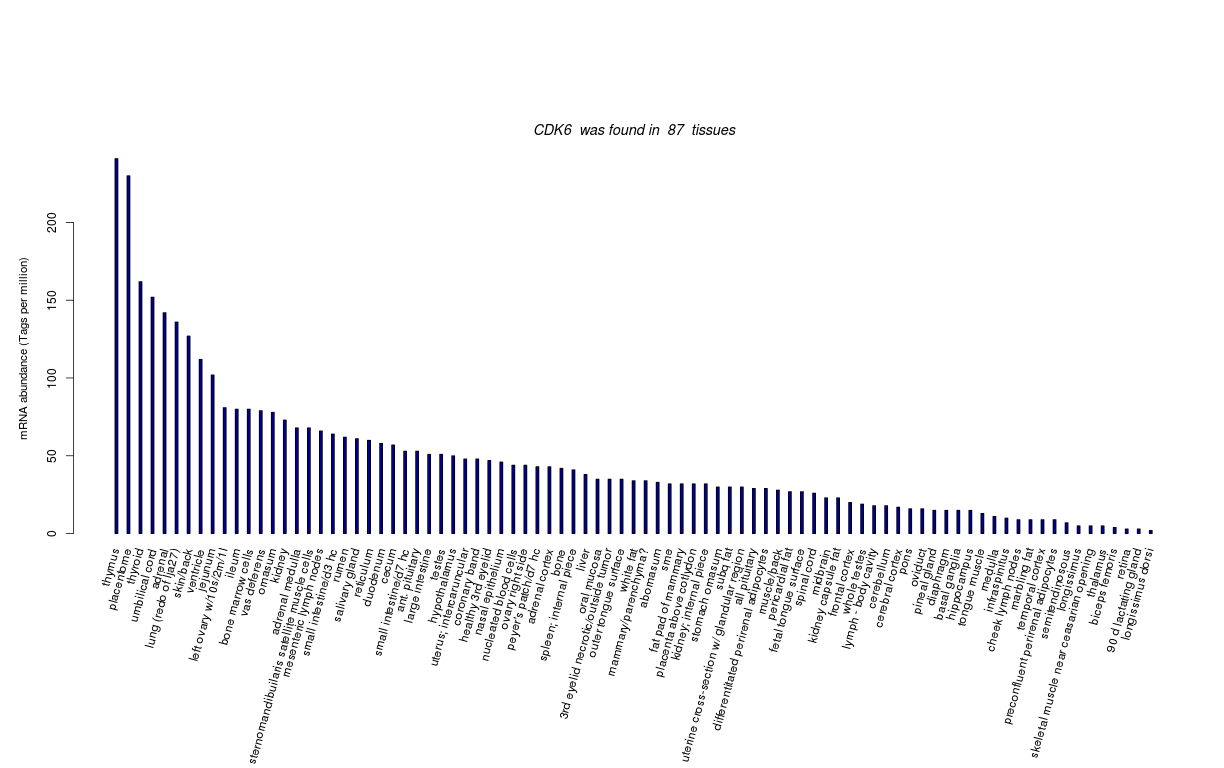
| Tag Count based mRNA-Abundances across 87 different Tissues (TPM).
Based on Data from Bovine Gene Atlas |
(Move your mouse over the image to view a more detailed version) |
||||||||||||||||||||||||||||||||