| Bos taurus Gene: BT.28580 | |||||||
|---|---|---|---|---|---|---|---|
| Summary | |||||||
| InnateDB Gene | IDBG-629185.3 | ||||||
| Last Modified | 2014-10-13 [Report errors or provide feedback] | ||||||
| Gene Symbol | BT.28580 | ||||||
| Gene Name | Uncharacterized protein | ||||||
| Synonyms | |||||||
| Species | Bos taurus | ||||||
| Ensembl Gene | ENSBTAG00000039307 | ||||||
| Encoded Proteins |
Uncharacterized protein
|
||||||
| Protein Structure | |||||||
| Useful resources | Stemformatics EHFPI ImmGen | ||||||
| Entrez Gene | |||||||
| Summary |
This gene does not have any Entrez summary - the following is the summary from its human ortholog ENSG00000137693:
This gene encodes the human ortholog of chicken YAP protein which binds to the SH3 domain of the Yes proto-oncogene product. This protein contains a WW domain that is found in various structural, regulatory and signaling molecules in yeast, nematode, and mammals, and may be involved in protein-protein interaction. [provided by RefSeq, Jul 2008] This gene encodes a downstream nuclear effector of the Hippo signaling pathway which is involved in development, growth, repair, and homeostasis. This gene is known to play a role in the development and progression of multiple cancers as a transcriptional regulator of this signaling pathway and may function as a potential target for cancer treatment. Alternative splicing results in multiple transcript variants encoding different isoforms. [provided by RefSeq, Aug 2013] |
||||||
| Gene Information | |||||||
| Type | Protein coding | ||||||
| Genomic Location | Chromosome 15:6750617-6817541 | ||||||
| Strand | Reverse strand | ||||||
| Band | |||||||
| Transcripts |
|
||||||
| Interactions | |||||||
| Number of Interactions |
This gene and/or its encoded proteins are associated with 0 experimentally validated interaction(s) in this database.
They are also associated with 82 interaction(s) predicted by orthology.
|
||||||
| Gene Ontology | |||||||
Molecular Function |
|
||||||
| Biological Process |
|
||||||
| Cellular Component |
|
||||||
| Orthologs | |||||||
|
Species
Homo sapiens
Mus musculus
|
Gene ID
Gene Order
|
||||||
| Pathway Predictions based on Human Orthology Data | |||||||
| NETPATH |
TGF_beta_Receptor pathway
|
||||||
| REACTOME |
Nuclear signaling by ERBB4 pathway
Signaling by ERBB4 pathway
Signaling by Hippo pathway
PPARA activates gene expression pathway
Regulation of lipid metabolism by Peroxisome proliferator-activated receptor alpha (PPARalpha) pathway
Fatty acid, triacylglycerol, and ketone body metabolism pathway
YAP1- and WWTR1 (TAZ)-stimulated gene expression pathway
Generic Transcription Pathway pathway
Metabolism of lipids and lipoproteins pathway
Signal Transduction pathway
Metabolism pathway
Gene Expression pathway
Nuclear signaling by ERBB4 pathway
Fatty acid, triacylglycerol, and ketone body metabolism pathway
Signaling by ERBB4 pathway
Metabolism pathway
Metabolism of lipids and lipoproteins pathway
Regulation of lipid metabolism by Peroxisome proliferator-activated receptor alpha (PPARalpha) pathway
Signaling by Hippo pathway
Gene Expression pathway
PPARA activates gene expression pathway
YAP1- and WWTR1 (TAZ)-stimulated gene expression pathway
Generic Transcription Pathway pathway
Signal Transduction pathway
|
||||||
| KEGG | |||||||
| INOH | |||||||
| PID NCI |
ErbB4 signaling events
p73 transcription factor network
TGF-beta receptor signaling
Validated transcriptional targets of deltaNp63 isoforms
|
||||||
| Cross-References | |||||||
| SwissProt | |||||||
| TrEMBL | F1MK73 | ||||||
| UniProt Splice Variant | |||||||
| Entrez Gene | 100336629 | ||||||
| UniGene | Bt.89292 | ||||||
| RefSeq | XM_003586931 XM_005215747 XM_005215749 | ||||||
| HUGO | |||||||
| OMIM | |||||||
| CCDS | |||||||
| HPRD | |||||||
| IMGT | |||||||
| EMBL | DAAA02039890 DAAA02039891 | ||||||
| GenPept | |||||||
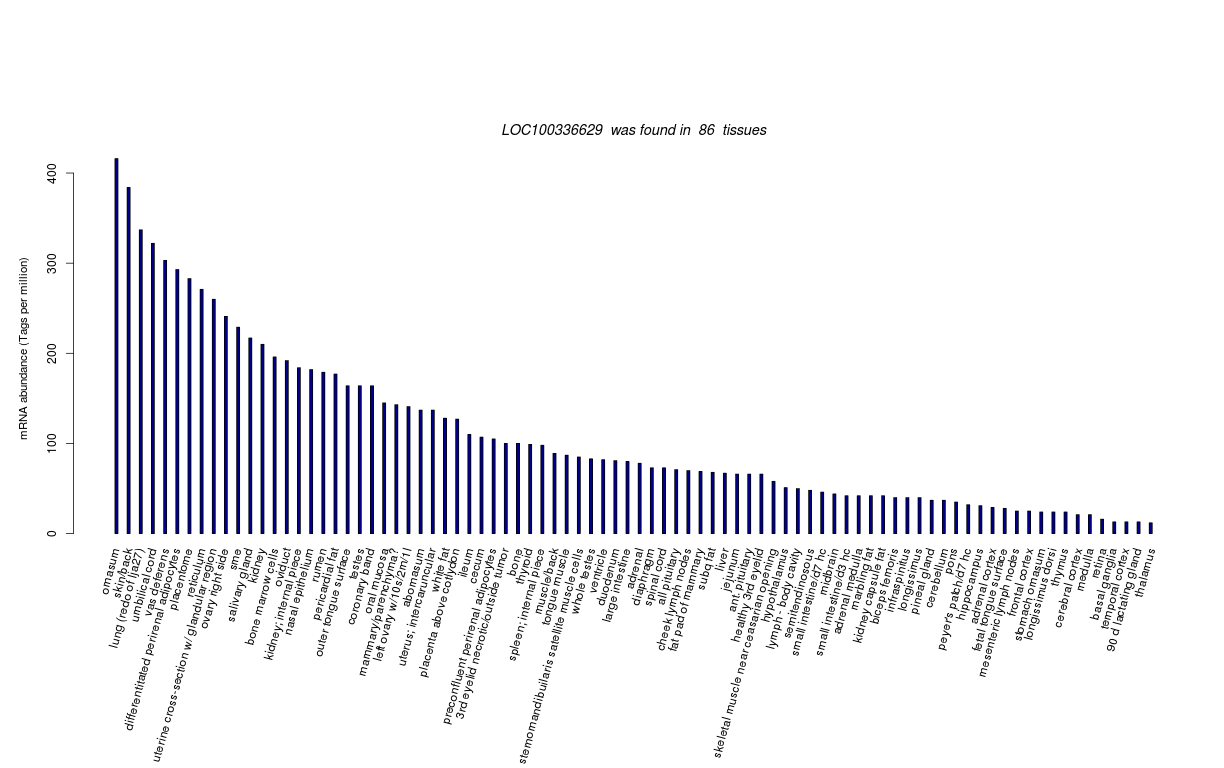
| RNA Seq Atlas | 100336629 | ||||||
| Transcript Frequencies | |||||||
| Tag Count based mRNA-Abundances across 87 different Tissues (TPM).
Based on Data from Bovine Gene Atlas |
(Move your mouse over the image to view a more detailed version) |
||||||