| Bos taurus Gene: XIAP | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Summary | |||||||||
| InnateDB Gene | IDBG-629983.3 | ||||||||
| Last Modified | 2014-10-13 [Report errors or provide feedback] | ||||||||
| Gene Symbol | XIAP | ||||||||
| Gene Name | baculoviral IAP repeat-containing protein 4 | ||||||||
| Synonyms | BIRC4 | ||||||||
| Species | Bos taurus | ||||||||
| Ensembl Gene | ENSBTAG00000040350 | ||||||||
| Encoded Proteins | |||||||||
| Protein Structure | |||||||||
| Useful resources | Stemformatics EHFPI ImmGen | ||||||||
| InnateDB Annotation from Orthologs | |||||||||
| Summary |
[Homo sapiens] XIAP regulates cytosol-specific innate immunity to Listeria infection.
[Homo sapiens] XIAP interacts with NOD1 and NOD2 and mediates NOD signalling via interaction with RIPK2.
[Homo sapiens] XIAP and its E3 ligase activity promote transforming growth factor-{beta}-mediated NF-kappaB activation during breast cancer progression.
[Homo sapiens] X-linked inhibitor of apoptosis protein (XIAP) in a complex with survivin, a physiological substrate for granzyme M (GzmM), act to inhibit caspase activation.
[Homo sapiens] XIAP facilitates ubiquitin-dependent signalling activated by pattern recognition receptors, such as TLR and NOD, to mediate the activation of NFKB transcription.
[Mus musculus] Xiap facilitates ubiquitin-dependent signalling activated by pattern recognition receptors, such as TLR and NOD, to mediate the activation of NFKB transcription.
|
||||||||
| Entrez Gene | |||||||||
| Summary |
This gene does not have any Entrez summary - the following is the summary from its human ortholog ENSG00000101966:
This gene encodes a protein that belongs to a family of apoptotic suppressor proteins. Members of this family share a conserved motif termed, baculovirus IAP repeat, which is necessary for their anti-apoptotic function. This protein functions through binding to tumor necrosis factor receptor-associated factors TRAF1 and TRAF2 and inhibits apoptosis induced by menadione, a potent inducer of free radicals, and interleukin 1-beta converting enzyme. This protein also inhibits at least two members of the caspase family of cell-death proteases, caspase-3 and caspase-7. Mutations in this gene are the cause of X-linked lymphoproliferative syndrome. Alternate splicing results in multiple transcript variants. Pseudogenes of this gene are found on chromosomes 2 and 11.[provided by RefSeq, Feb 2011] |
||||||||
| Gene Information | |||||||||
| Type | Protein coding | ||||||||
| Genomic Location | Chromosome X:7830563-7845821 | ||||||||
| Strand | Forward strand | ||||||||
| Band | |||||||||
| Transcripts |
|
||||||||
| Interactions | |||||||||
| Number of Interactions |
This gene and/or its encoded proteins are associated with 0 experimentally validated interaction(s) in this database.
They are also associated with 112 interaction(s) predicted by orthology.
|
||||||||
| Gene Ontology | |||||||||
Molecular Function |
|
||||||||
| Biological Process |
|
||||||||
| Cellular Component |
|
||||||||
| Orthologs | |||||||||
|
Species
Homo sapiens
Mus musculus
|
Gene ID
Gene Order
|
||||||||
| Pathway Predictions based on Human Orthology Data | |||||||||
| NETPATH |
TNFalpha pathway
|
||||||||
| REACTOME |
SMAC-mediated dissociation of IAP:caspase complexes pathway
SMAC binds to IAPs pathway
misspliced LRP5 mutants have enhanced beta-catenin-dependent signaling pathway
Signaling by WNT in cancer pathway
SMAC-mediated apoptotic response pathway
Signaling by Wnt pathway
deactivation of the beta-catenin transactivating complex pathway
Apoptosis pathway
Signal Transduction pathway
Apoptotic factor-mediated response pathway
TCF dependent signaling in response to WNT pathway
RNF mutants show enhanced WNT signaling and proliferation pathway
Intrinsic Pathway for Apoptosis pathway
XAV939 inhibits tankyrase, stabilizing AXIN pathway
Disease pathway
Apoptosis pathway
Disease pathway
SMAC-mediated dissociation of IAP:caspase complexes pathway
Signaling by Wnt pathway
Signaling by WNT in cancer pathway
SMAC-mediated apoptotic response pathway
deactivation of the beta-catenin transactivating complex pathway
misspliced LRP5 mutants have enhanced beta-catenin-dependent signaling pathway
RNF mutants show enhanced WNT signaling and proliferation pathway
TCF dependent signaling in response to WNT pathway
SMAC binds to IAPs pathway
Signal Transduction pathway
Apoptotic factor-mediated response pathway
XAV939 inhibits tankyrase, stabilizing AXIN pathway
Intrinsic Pathway for Apoptosis pathway
|
||||||||
| KEGG |
Apoptosis pathway
Ubiquitin mediated proteolysis pathway
Small cell lung cancer pathway
Focal adhesion pathway
Pathways in cancer pathway
Toxoplasmosis pathway
Ubiquitin mediated proteolysis pathway
Small cell lung cancer pathway
Apoptosis pathway
Focal adhesion pathway
Pathways in cancer pathway
Toxoplasmosis pathway
|
||||||||
| INOH |
TGF-beta signaling pathway
BMP2 signaling pathway
BMP2 signaling pathway
TGF-beta signaling pathway
|
||||||||
| PID NCI |
Caspase Cascade in Apoptosis
BMP receptor signaling
TGF-beta receptor signaling
|
||||||||
| Cross-References | |||||||||
| SwissProt | |||||||||
| TrEMBL | |||||||||
| UniProt Splice Variant | |||||||||
| Entrez Gene | |||||||||
| UniGene | Bt.13052 | ||||||||
| RefSeq | NM_001205592 | ||||||||
| HUGO | |||||||||
| OMIM | |||||||||
| CCDS | |||||||||
| HPRD | |||||||||
| IMGT | |||||||||
| EMBL | |||||||||
| GenPept | |||||||||
| RNA Seq Atlas | |||||||||
| Transcript Frequencies | |||||||||
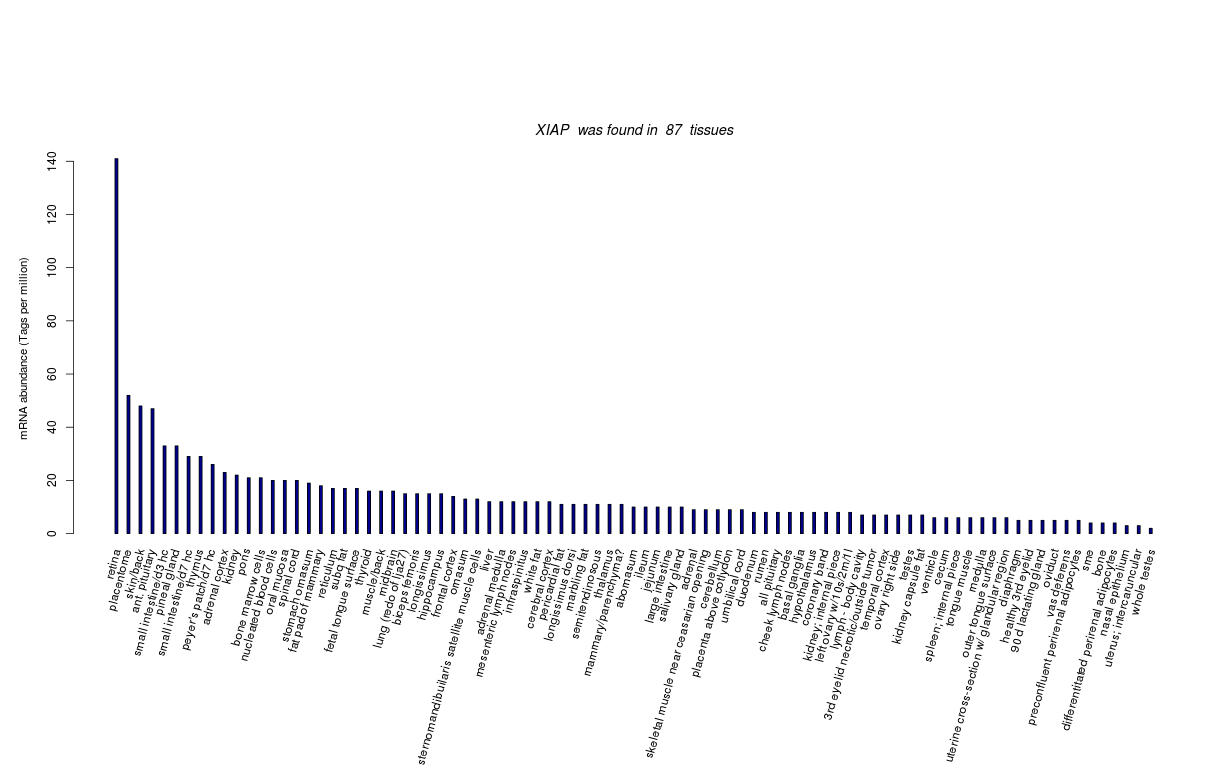
| Tag Count based mRNA-Abundances across 87 different Tissues (TPM).
Based on Data from Bovine Gene Atlas |
(Move your mouse over the image to view a more detailed version) |
||||||||