| Bos taurus Gene: PHGDH | |||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Summary | |||||||||||||||||||||||||||||||||||
| InnateDB Gene | IDBG-634247.3 | ||||||||||||||||||||||||||||||||||
| Last Modified | 2014-10-13 [Report errors or provide feedback] | ||||||||||||||||||||||||||||||||||
| Gene Symbol | PHGDH | ||||||||||||||||||||||||||||||||||
| Gene Name | D-3-phosphoglycerate dehydrogenase | ||||||||||||||||||||||||||||||||||
| Synonyms | |||||||||||||||||||||||||||||||||||
| Species | Bos taurus | ||||||||||||||||||||||||||||||||||
| Ensembl Gene | ENSBTAG00000039719 | ||||||||||||||||||||||||||||||||||
| Encoded Proteins |
D-3-phosphoglycerate dehydrogenase
|
||||||||||||||||||||||||||||||||||
| Protein Structure | |||||||||||||||||||||||||||||||||||
| Useful resources | Stemformatics EHFPI ImmGen | ||||||||||||||||||||||||||||||||||
| Entrez Gene | |||||||||||||||||||||||||||||||||||
| Summary |
This gene does not have any Entrez summary - the following is the summary from its human ortholog ENSG00000092621:
This gene encodes the enzyme which is involved in the early steps of L-serine synthesis in animal cells. L-serine is required for D-serine and other amino acid synthesis. The enzyme requires NAD/NADH as a cofactor and forms homotetramers for activity. Mutations in this gene have been found in a family with congenital microcephaly, psychomotor retardation and other symptoms. Multiple alternatively spliced transcript variants have been found, however the full-length nature of most are not known. [provided by RefSeq, Aug 2011] |
||||||||||||||||||||||||||||||||||
| Gene Information | |||||||||||||||||||||||||||||||||||
| Type | Protein coding | ||||||||||||||||||||||||||||||||||
| Genomic Location | Chromosome 3:23672339-23703529 | ||||||||||||||||||||||||||||||||||
| Strand | Reverse strand | ||||||||||||||||||||||||||||||||||
| Band | |||||||||||||||||||||||||||||||||||
| Transcripts |
|
||||||||||||||||||||||||||||||||||
| Interactions | |||||||||||||||||||||||||||||||||||
| Number of Interactions |
This gene and/or its encoded proteins are associated with 0 experimentally validated interaction(s) in this database.
They are also associated with 54 interaction(s) predicted by orthology.
|
||||||||||||||||||||||||||||||||||
| Gene Ontology | |||||||||||||||||||||||||||||||||||
Molecular Function |
|
||||||||||||||||||||||||||||||||||
| Biological Process |
|
||||||||||||||||||||||||||||||||||
| Cellular Component |
|
||||||||||||||||||||||||||||||||||
| Orthologs | |||||||||||||||||||||||||||||||||||
|
Species
Homo sapiens
Mus musculus
|
Gene ID
Gene Order
|
||||||||||||||||||||||||||||||||||
| Pathways | |||||||||||||||||||||||||||||||||||
| NETPATH | |||||||||||||||||||||||||||||||||||
| REACTOME |
Metabolism of amino acids and derivatives pathway
Serine biosynthesis pathway
Metabolism pathway
Amino acid synthesis and interconversion (transamination) pathway
|
||||||||||||||||||||||||||||||||||
| KEGG | |||||||||||||||||||||||||||||||||||
| INOH | |||||||||||||||||||||||||||||||||||
| PID NCI | |||||||||||||||||||||||||||||||||||
| Pathway Predictions based on Human Orthology Data | |||||||||||||||||||||||||||||||||||
| NETPATH | |||||||||||||||||||||||||||||||||||
| REACTOME |
Serine biosynthesis pathway
Amino acid synthesis and interconversion (transamination) pathway
Metabolism of amino acids and derivatives pathway
Metabolism pathway
Metabolism pathway
Serine biosynthesis pathway
Metabolism of amino acids and derivatives pathway
Amino acid synthesis and interconversion (transamination) pathway
|
||||||||||||||||||||||||||||||||||
| KEGG |
Glycine, serine and threonine metabolism pathway
Glycine, serine and threonine metabolism pathway
|
||||||||||||||||||||||||||||||||||
| INOH |
Glycine Serine metabolism pathway
|
||||||||||||||||||||||||||||||||||
| PID NCI | |||||||||||||||||||||||||||||||||||
| Cross-References | |||||||||||||||||||||||||||||||||||
| SwissProt | |||||||||||||||||||||||||||||||||||
| TrEMBL | |||||||||||||||||||||||||||||||||||
| UniProt Splice Variant | |||||||||||||||||||||||||||||||||||
| Entrez Gene | |||||||||||||||||||||||||||||||||||
| UniGene | Bt.53026 | ||||||||||||||||||||||||||||||||||
| RefSeq | NM_001035017 | ||||||||||||||||||||||||||||||||||
| HUGO | |||||||||||||||||||||||||||||||||||
| OMIM | |||||||||||||||||||||||||||||||||||
| CCDS | |||||||||||||||||||||||||||||||||||
| HPRD | |||||||||||||||||||||||||||||||||||
| IMGT | |||||||||||||||||||||||||||||||||||
| EMBL | |||||||||||||||||||||||||||||||||||
| GenPept | |||||||||||||||||||||||||||||||||||
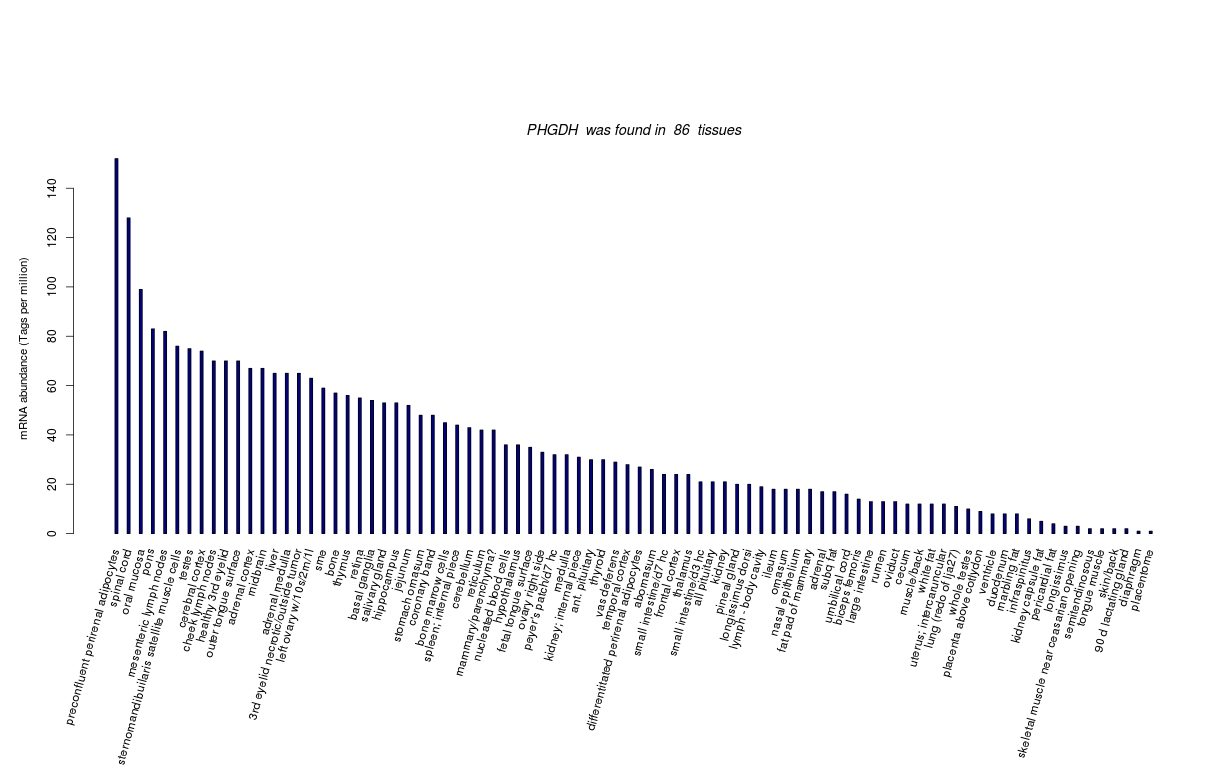
| RNA Seq Atlas | |||||||||||||||||||||||||||||||||||
| Transcript Frequencies | |||||||||||||||||||||||||||||||||||
| Tag Count based mRNA-Abundances across 87 different Tissues (TPM).
Based on Data from Bovine Gene Atlas |
(Move your mouse over the image to view a more detailed version) |
||||||||||||||||||||||||||||||||||