| Bos taurus Gene: CDS1 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Summary | |||||||||||
| InnateDB Gene | IDBG-643256.3 | ||||||||||
| Last Modified | 2014-10-13 [Report errors or provide feedback] | ||||||||||
| Gene Symbol | CDS1 | ||||||||||
| Gene Name | Phosphatidate cytidylyltransferase | ||||||||||
| Synonyms | |||||||||||
| Species | Bos taurus | ||||||||||
| Ensembl Gene | ENSBTAG00000045787 | ||||||||||
| Encoded Proteins |
CDP-diacylglycerol synthase (phosphatidate cytidylyltransferase) 1
|
||||||||||
| Protein Structure | |||||||||||
| Useful resources | Stemformatics EHFPI ImmGen | ||||||||||
| Entrez Gene | |||||||||||
| Summary |
This gene does not have any Entrez summary - the following is the summary from its human ortholog ENSG00000163624:
Breakdown products of phosphoinositides are ubiquitous second messengers that function downstream of many G protein-coupled receptors and tyrosine kinases regulating cell growth, calcium metabolism, and protein kinase C activity. This gene encodes an enzyme which regulates the amount of phosphatidylinositol available for signaling by catalyzing the conversion of phosphatidic acid to CDP-diacylglycerol. This enzyme is an integral membrane protein localized to two subcellular domains, the matrix side of the inner mitochondrial membrane where it is thought to be involved in the synthesis of phosphatidylglycerol and cardiolipin and the cytoplasmic side of the endoplasmic reticulum where it functions in phosphatidylinositol biosynthesis. Two genes encoding this enzyme have been identified in humans, one mapping to human chromosome 4q21 and a second to 20p13. [provided by RefSeq, Jul 2008] |
||||||||||
| Gene Information | |||||||||||
| Type | Protein coding | ||||||||||
| Genomic Location | Chromosome 6:101195831-101270493 | ||||||||||
| Strand | Forward strand | ||||||||||
| Band | |||||||||||
| Transcripts |
|
||||||||||
| Interactions | |||||||||||
| Number of Interactions |
This gene and/or its encoded proteins are associated with 0 experimentally validated interaction(s) in this database.
They are also associated with 1 interaction(s) predicted by orthology.
|
||||||||||
| Gene Ontology | |||||||||||
Molecular Function |
|
||||||||||
| Biological Process |
|
||||||||||
| Cellular Component |
|
||||||||||
| Orthologs | |||||||||||
|
Species
Homo sapiens
Mus musculus
|
Gene ID
Gene Order
|
||||||||||
| Pathway Predictions based on Human Orthology Data | |||||||||||
| NETPATH | |||||||||||
| REACTOME |
Synthesis of PI pathway
Metabolism of lipids and lipoproteins pathway
Glycerophospholipid biosynthesis pathway
Phospholipid metabolism pathway
Metabolism pathway
Synthesis of PI pathway
Metabolism pathway
Metabolism of lipids and lipoproteins pathway
Glycerophospholipid biosynthesis pathway
Phospholipid metabolism pathway
|
||||||||||
| KEGG |
Glycerophospholipid metabolism pathway
Phosphatidylinositol signaling system pathway
Glycerophospholipid metabolism pathway
Phosphatidylinositol signaling system pathway
|
||||||||||
| INOH | |||||||||||
| PID NCI | |||||||||||
| Cross-References | |||||||||||
| SwissProt | |||||||||||
| TrEMBL | G3MZU2 | ||||||||||
| UniProt Splice Variant | |||||||||||
| Entrez Gene | 100336936 | ||||||||||
| UniGene | |||||||||||
| RefSeq | XM_003586222 | ||||||||||
| HUGO | HGNC:1800 | ||||||||||
| OMIM | |||||||||||
| CCDS | |||||||||||
| HPRD | |||||||||||
| IMGT | |||||||||||
| EMBL | DAAA02018308 DAAA02018309 DAAA02018310 DAAA02018311 | ||||||||||
| GenPept | |||||||||||
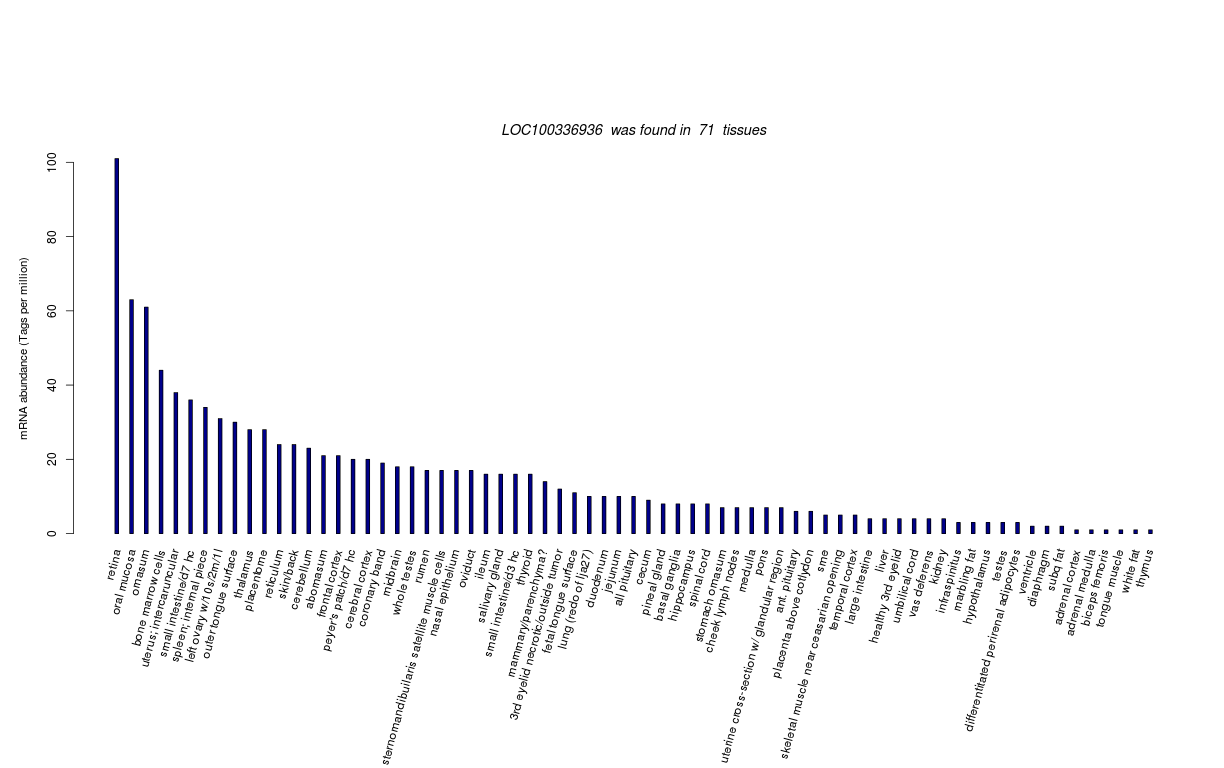
| RNA Seq Atlas | 100336936 | ||||||||||
| Transcript Frequencies | |||||||||||
| Tag Count based mRNA-Abundances across 87 different Tissues (TPM).
Based on Data from Bovine Gene Atlas |
(Move your mouse over the image to view a more detailed version) |
||||||||||