| Bos taurus Gene: POLR2H | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Summary | |||||||||
| InnateDB Gene | IDBG-635056.3 | ||||||||
| Last Modified | 2014-10-13 [Report errors or provide feedback] | ||||||||
| Gene Symbol | POLR2H | ||||||||
| Gene Name | DNA-directed RNA polymerases I, II, and III subunit RPABC3 | ||||||||
| Synonyms | |||||||||
| Species | Bos taurus | ||||||||
| Ensembl Gene | ENSBTAG00000040199 | ||||||||
| Encoded Proteins |
polymerase (RNA) II (DNA directed) polypeptide H
|
||||||||
| Protein Structure | |||||||||
| Useful resources | Stemformatics EHFPI ImmGen | ||||||||
| Entrez Gene | |||||||||
| Summary |
This gene does not have any Entrez summary - the following is the summary from its human ortholog ENSG00000163882:
This gene encodes one of the essential subunits of RNA polymerase II that is shared by the other two eukaryotic DNA-directed RNA polymerases, I and III. [provided by RefSeq, Jul 2008] The three eukaryotic RNA polymerases are complex multisubunit enzymes that play a central role in the transcription of nuclear genes. This gene encodes an essential and highly conserved subunit of RNA polymerase II that is shared by the other two eukaryotic DNA-directed RNA polymerases, I and III. Alternative splicing results in multiple transcript variants of this gene. [provided by RefSeq, Jul 2013] |
||||||||
| Gene Information | |||||||||
| Type | Protein coding | ||||||||
| Genomic Location | Chromosome 1:83437865-83442294 | ||||||||
| Strand | Reverse strand | ||||||||
| Band | |||||||||
| Transcripts |
|
||||||||
| Interactions | |||||||||
| Number of Interactions |
This gene and/or its encoded proteins are associated with 0 experimentally validated interaction(s) in this database.
They are also associated with 41 interaction(s) predicted by orthology.
|
||||||||
| Gene Ontology | |||||||||
Molecular Function |
|
||||||||
| Biological Process |
|
||||||||
| Cellular Component |
|
||||||||
| Orthologs | |||||||||
|
Species
Homo sapiens
Mus musculus
|
Gene ID
Gene Order
|
||||||||
| Pathway Predictions based on Human Orthology Data | |||||||||
| NETPATH |
TNFalpha pathway
|
||||||||
| REACTOME |
Tat-mediated HIV elongation arrest and recovery pathway
Pausing and recovery of Tat-mediated HIV elongation pathway
Viral Messenger RNA Synthesis pathway
RNA Polymerase III Abortive And Retractive Initiation pathway
RNA Polymerase III Transcription Termination pathway
RNA Polymerase III Chain Elongation pathway
RNA Polymerase III Transcription Initiation From Type 3 Promoter pathway
RNA Polymerase III Transcription Initiation From Type 2 Promoter pathway
RNA Polymerase III Transcription Initiation From Type 1 Promoter pathway
RNA Polymerase I Transcription Termination pathway
RNA Polymerase I Chain Elongation pathway
RNA Polymerase I Promoter Escape pathway
RNA Polymerase I Transcription Initiation pathway
mRNA Splicing - Major Pathway pathway
mRNA Splicing - Minor Pathway pathway
Processing of Capped Intron-Containing Pre-mRNA pathway
mRNA Capping pathway
MicroRNA (miRNA) biogenesis pathway
RNA Polymerase II Transcription Pre-Initiation And Promoter Opening pathway
RNA Polymerase II Pre-transcription Events pathway
Formation of RNA Pol II elongation complex pathway
Formation of the Early Elongation Complex pathway
RNA Polymerase II Transcription Elongation pathway
RNA Polymerase II Promoter Escape pathway
RNA Polymerase II Transcription Initiation pathway
RNA Polymerase II Transcription Initiation And Promoter Clearance pathway
RNA Pol II CTD phosphorylation and interaction with CE pathway
Formation of HIV elongation complex in the absence of HIV Tat pathway
HIV Transcription Initiation pathway
Pausing and recovery of HIV elongation pathway
RNA Polymerase II HIV Promoter Escape pathway
Formation of the HIV-1 Early Elongation Complex pathway
Formation of HIV-1 elongation complex containing HIV-1 Tat pathway
Tat-mediated elongation of the HIV-1 transcript pathway
Abortive elongation of HIV-1 transcript in the absence of Tat pathway
RNA Pol II CTD phosphorylation and interaction with CE pathway
HIV elongation arrest and recovery pathway
Transcription of the HIV genome pathway
Late Phase of HIV Life Cycle pathway
Formation of transcription-coupled NER (TC-NER) repair complex pathway
Dual incision reaction in TC-NER pathway
Transcription-coupled NER (TC-NER) pathway
Influenza Infection pathway
RNA Polymerase I Transcription pathway
RNA Polymerase III Transcription Initiation pathway
Innate Immune System pathway
Transcriptional regulation by small RNAs pathway
Epigenetic regulation of gene expression pathway
Influenza Viral RNA Transcription and Replication pathway
RNA Polymerase I Promoter Clearance pathway
NoRC negatively regulates rRNA expression pathway
HIV Transcription Elongation pathway
RNA Polymerase II Transcription pathway
Immune System pathway
HIV Life Cycle pathway
RNA Polymerase III Transcription pathway
HIV Infection pathway
RNA Polymerase I, RNA Polymerase III, and Mitochondrial Transcription pathway
Influenza Life Cycle pathway
mRNA Splicing pathway
Cytosolic sensors of pathogen-associated DNA pathway
Nucleotide Excision Repair pathway
Gene Expression pathway
Disease pathway
DNA Repair pathway
Negative epigenetic regulation of rRNA expression pathway
Regulatory RNA pathways pathway
RNA Polymerase II Transcription Initiation And Promoter Clearance pathway
Transcription-coupled NER (TC-NER) pathway
Innate Immune System pathway
RNA Polymerase I Transcription pathway
RNA Polymerase I, RNA Polymerase III, and Mitochondrial Transcription pathway
Formation of the Early Elongation Complex pathway
mRNA Splicing pathway
Formation of transcription-coupled NER (TC-NER) repair complex pathway
RNA Polymerase III Transcription Initiation pathway
RNA Polymerase II Transcription Initiation pathway
mRNA Splicing - Minor Pathway pathway
RNA Polymerase II Promoter Escape pathway
RNA Polymerase III Transcription Initiation From Type 1 Promoter pathway
RNA Polymerase I Transcription Termination pathway
Immune System pathway
Nucleotide Excision Repair pathway
Formation of RNA Pol II elongation complex pathway
RNA Polymerase II Transcription Pre-Initiation And Promoter Opening pathway
RNA Polymerase III Transcription Initiation From Type 2 Promoter pathway
RNA Polymerase III Abortive And Retractive Initiation pathway
DNA Repair pathway
Transcriptional regulation by small RNAs pathway
RNA Polymerase II Transcription Elongation pathway
RNA Polymerase II Pre-transcription Events pathway
Cytosolic sensors of pathogen-associated DNA pathway
mRNA Capping pathway
Gene Expression pathway
Regulatory RNA pathways pathway
RNA Polymerase III Transcription Termination pathway
mRNA Splicing - Major Pathway pathway
RNA Polymerase I Promoter Escape pathway
Dual incision reaction in TC-NER pathway
RNA Polymerase I Promoter Clearance pathway
RNA Polymerase I Chain Elongation pathway
Processing of Capped Intron-Containing Pre-mRNA pathway
RNA Polymerase II Transcription pathway
RNA Polymerase III Transcription pathway
RNA Pol II CTD phosphorylation and interaction with CE pathway
RNA Polymerase III Transcription Initiation From Type 3 Promoter pathway
RNA Polymerase I Transcription Initiation pathway
RNA Polymerase III Chain Elongation pathway
|
||||||||
| KEGG |
Pyrimidine metabolism pathway
Purine metabolism pathway
RNA polymerase pathway
Huntington's disease pathway
Cytosolic DNA-sensing pathway pathway
Pyrimidine metabolism pathway
RNA polymerase pathway
Purine metabolism pathway
Huntington's disease pathway
Cytosolic DNA-sensing pathway pathway
|
||||||||
| INOH | |||||||||
| PID NCI | |||||||||
| Cross-References | |||||||||
| SwissProt | |||||||||
| TrEMBL | A8QQK8 F2Z4H3 | ||||||||
| UniProt Splice Variant | |||||||||
| Entrez Gene | 505599 | ||||||||
| UniGene | Bt.106863 | ||||||||
| RefSeq | NM_001192065 | ||||||||
| HUGO | |||||||||
| OMIM | |||||||||
| CCDS | |||||||||
| HPRD | |||||||||
| IMGT | |||||||||
| EMBL | BC118247 DAAA02001883 | ||||||||
| GenPept | AAI18248 | ||||||||
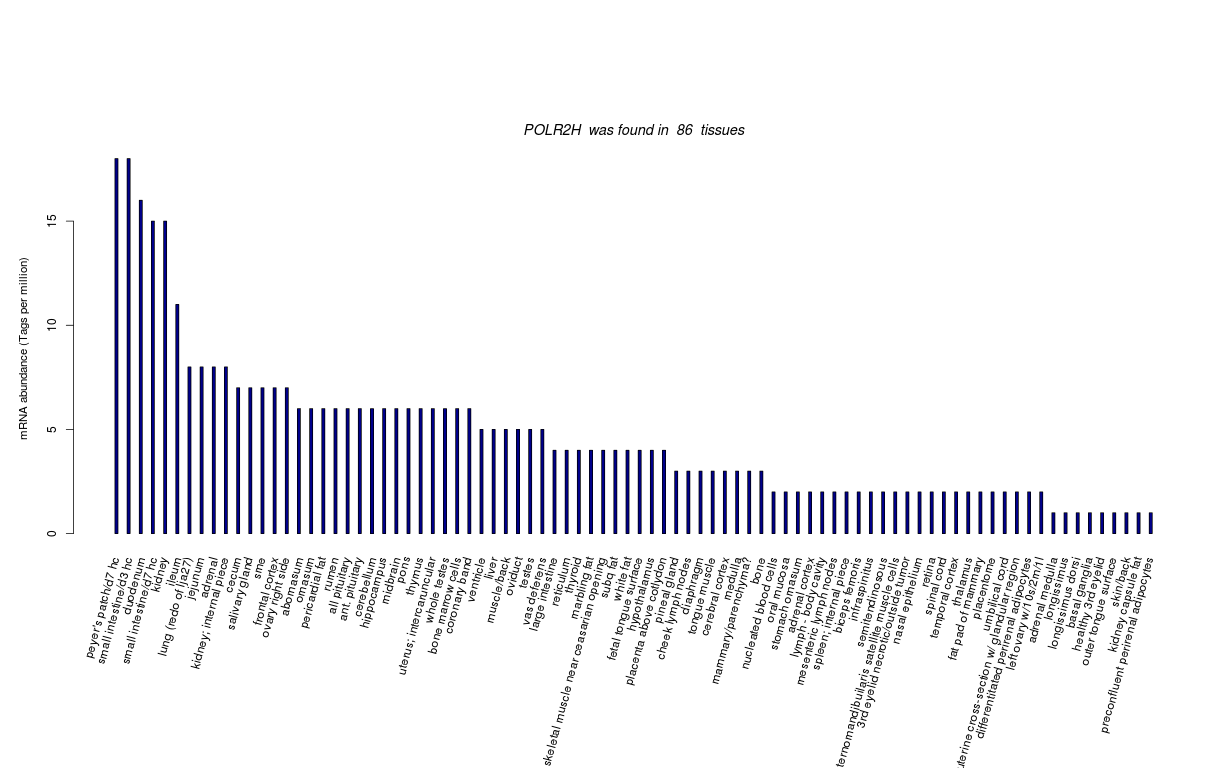
| RNA Seq Atlas | 505599 | ||||||||
| Transcript Frequencies | |||||||||
| Tag Count based mRNA-Abundances across 87 different Tissues (TPM).
Based on Data from Bovine Gene Atlas |
(Move your mouse over the image to view a more detailed version) |
||||||||