| Bos taurus Gene: WT1 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Summary | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| InnateDB Gene | IDBG-636230.3 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Last Modified | 2014-10-13 [Report errors or provide feedback] | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Gene Symbol | WT1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Gene Name | Uncharacterized protein | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Synonyms | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Species | Bos taurus | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Ensembl Gene | ENSBTAG00000047268 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Encoded Proteins |
Wilms tumor 1
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Protein Structure | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Useful resources | Stemformatics EHFPI ImmGen | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Entrez Gene | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Summary |
This gene does not have any Entrez summary - the following is the summary from its human ortholog ENSG00000184937:
This gene encodes a transcription factor that contains four zinc-finger motifs at the C-terminus and a proline/glutamine-rich DNA-binding domain at the N-terminus. It has an essential role in the normal development of the urogenital system, and it is mutated in a small subset of patients with Wilm's tumors. This gene exhibits complex tissue-specific and polymorphic imprinting pattern, with biallelic, and monoallelic expression from the maternal and paternal alleles in different tissues. Multiple transcript variants have been described. In several variants, there is evidence for the use of a non-AUG (CUG) translation initiation site upstream of and in-frame with the first AUG. Authors of PMID:7926762 also provide evidence that WT1 mRNA undergoes RNA editing in human and rat, and that this process is tissue-restricted and developmentally regulated. [provided by RefSeq, Oct 2010] This gene encodes a transcription factor that contains four zinc-finger motifs at the C-terminus and a proline/glutamine-rich DNA-binding domain at the N-terminus. It has an essential role in the normal development of the urogenital system, and it is mutated in a small subset of patients with Wilm\'s tumors. This gene exhibits complex tissue-specific and polymorphic imprinting pattern, with biallelic, and monoallelic expression from the maternal and paternal alleles in different tissues. Multiple transcript variants have been described. In several variants, there is evidence for the use of a non-AUG (CUG) translation initiation site upstream of and in-frame with the first AUG. Authors of PMID:7926762 also provide evidence that WT1 mRNA undergoes RNA editing in human and rat, and that this process is tissue-restricted and developmentally regulated. [provided by RefSeq, Oct 2010] |
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Gene Information | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Type | Protein coding | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Genomic Location | Chromosome 15:63913734-63965294 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Strand | Reverse strand | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Band | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Transcripts |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Interactions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Number of Interactions |
This gene and/or its encoded proteins are associated with 0 experimentally validated interaction(s) in this database.
They are also associated with 20 interaction(s) predicted by orthology.
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Gene Ontology | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Molecular Function |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Biological Process |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Cellular Component |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Orthologs | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
Species
Homo sapiens
Mus musculus
|
Gene ID
Gene Order
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Pathway Predictions based on Human Orthology Data | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| NETPATH | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| REACTOME | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| KEGG | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| INOH |
TGF-beta signaling pathway
BMP2 signaling pathway
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| PID NCI |
p73 transcription factor network
Regulation of Telomerase
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Cross-References | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| SwissProt | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| TrEMBL | G3MWM8 Q9TT81 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| UniProt Splice Variant | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Entrez Gene | 782183 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| UniGene | Bt.65911 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| RefSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| HUGO | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| OMIM | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| CCDS | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| HPRD | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| IMGT | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| EMBL | AF201738 DAAA02041238 DAAA02041239 DAAA02041240 DAAA02041241 DAAA02041242 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| GenPept | AAF19824 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| RNA Seq Atlas | 782183 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Transcript Frequencies | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
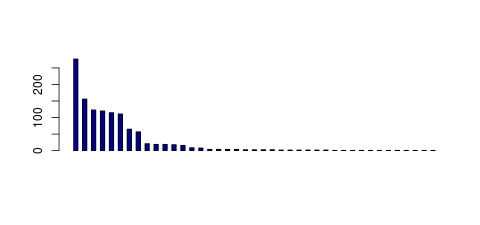
| Tag Count based mRNA-Abundances across 87 different Tissues (TPM).
Based on Data from Bovine Gene Atlas |
(Move your mouse over the image to view a more detailed version) |
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||