| Bos taurus Gene: TAF4 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Summary | |||||||||||
| InnateDB Gene | IDBG-641028.3 | ||||||||||
| Last Modified | 2014-10-13 [Report errors or provide feedback] | ||||||||||
| Gene Symbol | TAF4 | ||||||||||
| Gene Name | Uncharacterized protein | ||||||||||
| Synonyms | |||||||||||
| Species | Bos taurus | ||||||||||
| Ensembl Gene | ENSBTAG00000047991 | ||||||||||
| Encoded Proteins |
TAF4 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 135kDa
TAF4 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 135kDa
|
||||||||||
| Protein Structure | |||||||||||
| Useful resources | Stemformatics EHFPI ImmGen | ||||||||||
| Entrez Gene | |||||||||||
| Summary |
This gene does not have any Entrez summary - the following is the summary from its human ortholog ENSG00000130699:
microRNAs (miRNAs) are short (20-24 nt) non-coding RNAs that are involved in post-transcriptional regulation of gene expression in multicellular organisms by affecting both the stability and translation of mRNAs. miRNAs are transcribed by RNA polymerase II as part of capped and polyadenylated primary transcripts (pri-miRNAs) that can be either protein-coding or non-coding. The primary transcript is cleaved by the Drosha ribonuclease III enzyme to produce an approximately 70-nt stem-loop precursor miRNA (pre-miRNA), which is further cleaved by the cytoplasmic Dicer ribonuclease to generate the mature miRNA and antisense miRNA star (miRNA*) products. The mature miRNA is incorporated into a RNA-induced silencing complex (RISC), which recognizes target mRNAs through imperfect base pairing with the miRNA and most commonly results in translational inhibition or destabilization of the target mRNA. The RefSeq represents the predicted microRNA stem-loop. [provided by RefSeq, Sep 2009] Initiation of transcription by RNA polymerase II requires the activities of more than 70 polypeptides. The protein that coordinates these activities is transcription factor IID (TFIID), which binds to the core promoter to position the polymerase properly, serves as the scaffold for assembly of the remainder of the transcription complex, and acts as a channel for regulatory signals. TFIID is composed of the TATA-binding protein (TBP) and a group of evolutionarily conserved proteins known as TBP-associated factors or TAFs. TAFs may participate in basal transcription, serve as coactivators, function in promoter recognition or modify general transcription factors (GTFs) to facilitate complex assembly and transcription initiation. This gene encodes one of the larger subunits of TFIID that has been shown to potentiate transcriptional activation by retinoic acid, thyroid hormone and vitamin D3 receptors. In addition, this subunit interacts with the transcription factor CREB, which has a glutamine-rich activation domain, and binds to other proteins containing glutamine-rich regions. Aberrant binding to this subunit by proteins with expanded polyglutamine regions has been suggested as one of the pathogenetic mechanisms underlying a group of neurodegenerative disorders referred to as polyglutamine diseases. [provided by RefSeq, Jul 2008] Initiation of transcription by RNA polymerase II requires the activities of more than 70 polypeptides. The protein that coordinates these activities is transcription factor IID (TFIID), which binds to the core promoter to position the polymerase properly, serves as the scaffold for assembly of the remainder of the transcription complex, and acts as a channel for regulatory signals. TFIID is composed of the TATA-binding protein (TBP) and a group of evolutionarily conserved proteins known as TBP-associated factors or TAFs. TAFs may participate in basal transcription, serve as coactivators, function in promoter recognition or modify general transcription factors (GTFs) to facilitate complex assembly and transcription initiation. This gene encodes one of the larger subunits of TFIID that has been shown to potentiate transcriptional activation by retinoic acid, thyroid hormone and vitamin D3 receptors. In addition, this subunit interacts with the transcription factor CREB, which has a glutamine-rich activation domain, and binds to other proteins containing glutamine-rich regions. Aberrant binding to this subunit by proteins with expanded polyglutamine regions has been suggested as one of the pathogenetic mechanisms underlying a group of neurodegenerative disorders referred to as polyglutamine diseases. [provided by RefSeq, Jul 2008] |
||||||||||
| Gene Information | |||||||||||
| Type | Protein coding | ||||||||||
| Genomic Location | Chromosome 13:55627852-55655516 | ||||||||||
| Strand | Forward strand | ||||||||||
| Band | |||||||||||
| Transcripts |
|
||||||||||
| Interactions | |||||||||||
| Number of Interactions |
This gene and/or its encoded proteins are associated with 0 experimentally validated interaction(s) in this database.
They are also associated with 30 interaction(s) predicted by orthology.
|
||||||||||
| Gene Ontology | |||||||||||
Molecular Function |
|
||||||||||
| Biological Process |
|
||||||||||
| Cellular Component |
|
||||||||||
| Orthologs | |||||||||||
|
Species
Homo sapiens
Mus musculus
|
Gene ID
Gene Order
|
||||||||||
| Pathway Predictions based on Human Orthology Data | |||||||||||
| NETPATH | |||||||||||
| REACTOME |
RNA Polymerase II Transcription Pre-Initiation And Promoter Opening pathway
RNA Polymerase II Pre-transcription Events pathway
RNA Polymerase II Promoter Escape pathway
RNA Polymerase II Transcription Initiation pathway
RNA Polymerase II Transcription Initiation And Promoter Clearance pathway
HIV Transcription Initiation pathway
RNA Polymerase II HIV Promoter Escape pathway
Transcription of the HIV genome pathway
Late Phase of HIV Life Cycle pathway
RNA Polymerase II Transcription pathway
HIV Life Cycle pathway
HIV Infection pathway
Gene Expression pathway
Disease pathway
|
||||||||||
| KEGG |
Basal transcription factors pathway
Huntington's disease pathway
Basal transcription factors pathway
Huntington's disease pathway
|
||||||||||
| INOH | |||||||||||
| PID NCI | |||||||||||
| Cross-References | |||||||||||
| SwissProt | |||||||||||
| TrEMBL | F1MMS1 | ||||||||||
| UniProt Splice Variant | |||||||||||
| Entrez Gene | 789854 | ||||||||||
| UniGene | Bt.111113 | ||||||||||
| RefSeq | XM_001787927 XM_002692259 | ||||||||||
| HUGO | HGNC:11537 | ||||||||||
| OMIM | |||||||||||
| CCDS | |||||||||||
| HPRD | |||||||||||
| IMGT | |||||||||||
| EMBL | DAAA02036266 DAAA02036267 | ||||||||||
| GenPept | |||||||||||
| RNA Seq Atlas | 789854 | ||||||||||
| Transcript Frequencies | |||||||||||
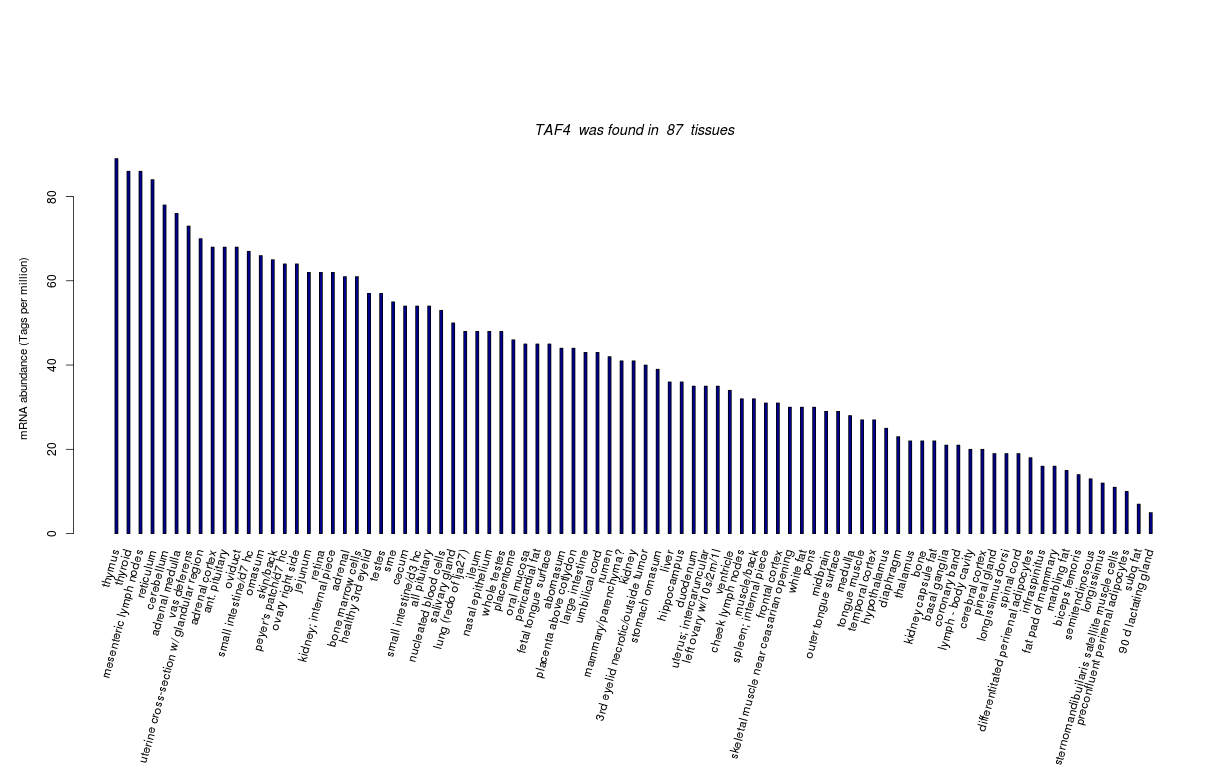
| Tag Count based mRNA-Abundances across 87 different Tissues (TPM).
Based on Data from Bovine Gene Atlas |
(Move your mouse over the image to view a more detailed version) |
||||||||||