| Bos taurus Gene: CXCL9 | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Summary | |||||||||||||||||||
| InnateDB Gene | IDBG-643104.3 | ||||||||||||||||||
| Last Modified | 2014-10-13 [Report errors or provide feedback] | ||||||||||||||||||
| Gene Symbol | CXCL9 | ||||||||||||||||||
| Gene Name | C-X-C motif chemokine 9 | ||||||||||||||||||
| Synonyms | |||||||||||||||||||
| Species | Bos taurus | ||||||||||||||||||
| Ensembl Gene | ENSBTAG00000038639 | ||||||||||||||||||
| Encoded Proteins |
C-X-C motif chemokine 9
|
||||||||||||||||||
| Protein Structure | |||||||||||||||||||
| Useful resources | Stemformatics EHFPI ImmGen | ||||||||||||||||||
| InnateDB Annotation from Orthologs | |||||||||||||||||||
| Summary |
[Homo sapiens] CXCL9 exert direct antimicrobial effects in vitro against Bacillus anthracis spore and bacilli in a receptor-independent manner and contributes to pulmonary innate immunity. (Demonstrated in murine model)
[Mus musculus] Cxcl9 exert direct antimicrobial effects in vitro against Bacillus anthracis spores and bacilli in a receptor-independent manner and contributes to pulmonary innate immunity.
|
||||||||||||||||||
| Entrez Gene | |||||||||||||||||||
| Summary |
This gene does not have any Entrez summary - the following is the summary from its human ortholog ENSG00000138755:
The function of this gene has not been specifically defined; however, it is thought to be involved in T cell trafficking. This gene has been localized to 4q21 with INP10, which is also a member of the chemokine family of cytokines. [provided by RefSeq, Jul 2008] This antimicrobial gene encodes a protein thought to be involved in T cell trafficking. The encoded protein binds to C-X-C motif chemokine 3 and is a chemoattractant for lymphocytes but not for neutrophils. [provided by RefSeq, Sep 2014] |
||||||||||||||||||
| Gene Information | |||||||||||||||||||
| Type | Protein coding | ||||||||||||||||||
| Genomic Location | Chromosome 6:92600177-92604818 | ||||||||||||||||||
| Strand | Reverse strand | ||||||||||||||||||
| Band | |||||||||||||||||||
| Transcripts |
|
||||||||||||||||||
| Interactions | |||||||||||||||||||
| Number of Interactions |
This gene and/or its encoded proteins are associated with 0 experimentally validated interaction(s) in this database.
They are also associated with 11 interaction(s) predicted by orthology.
|
||||||||||||||||||
| Gene Ontology | |||||||||||||||||||
Molecular Function |
|
||||||||||||||||||
| Biological Process |
|
||||||||||||||||||
| Cellular Component |
|
||||||||||||||||||
| Orthologs | |||||||||||||||||||
|
Species
Homo sapiens
Mus musculus
|
Gene ID
Gene Order
|
||||||||||||||||||
| Pathways | |||||||||||||||||||
| NETPATH | |||||||||||||||||||
| REACTOME |
Peptide ligand-binding receptors pathway
Signaling by GPCR pathway
G alpha (i) signalling events pathway
Signal Transduction pathway
GPCR downstream signaling pathway
GPCR ligand binding pathway
Class A/1 (Rhodopsin-like receptors) pathway
Chemokine receptors bind chemokines pathway
|
||||||||||||||||||
| KEGG | |||||||||||||||||||
| INOH | |||||||||||||||||||
| PID NCI | |||||||||||||||||||
| Pathway Predictions based on Human Orthology Data | |||||||||||||||||||
| NETPATH | |||||||||||||||||||
| REACTOME |
G alpha (i) signalling events pathway
Chemokine receptors bind chemokines pathway
Peptide ligand-binding receptors pathway
Class A/1 (Rhodopsin-like receptors) pathway
Signaling by GPCR pathway
Signal Transduction pathway
GPCR downstream signaling pathway
GPCR ligand binding pathway
GPCR downstream signaling pathway
Signaling by GPCR pathway
Class A/1 (Rhodopsin-like receptors) pathway
GPCR ligand binding pathway
Peptide ligand-binding receptors pathway
G alpha (i) signalling events pathway
Chemokine receptors bind chemokines pathway
Signal Transduction pathway
|
||||||||||||||||||
| KEGG |
Cytokine-cytokine receptor interaction pathway
Toll-like receptor signaling pathway pathway
Chemokine signaling pathway pathway
Cytokine-cytokine receptor interaction pathway
Toll-like receptor signaling pathway pathway
Chemokine signaling pathway pathway
|
||||||||||||||||||
| INOH | |||||||||||||||||||
| PID NCI |
IL23-mediated signaling events
CXCR3-mediated signaling events
|
||||||||||||||||||
| Cross-References | |||||||||||||||||||
| SwissProt | |||||||||||||||||||
| TrEMBL | |||||||||||||||||||
| UniProt Splice Variant | |||||||||||||||||||
| Entrez Gene | |||||||||||||||||||
| UniGene | |||||||||||||||||||
| RefSeq | NM_001113172 | ||||||||||||||||||
| HUGO | |||||||||||||||||||
| OMIM | |||||||||||||||||||
| CCDS | |||||||||||||||||||
| HPRD | |||||||||||||||||||
| IMGT | |||||||||||||||||||
| EMBL | |||||||||||||||||||
| GenPept | |||||||||||||||||||
| RNA Seq Atlas | |||||||||||||||||||
| Transcript Frequencies | |||||||||||||||||||
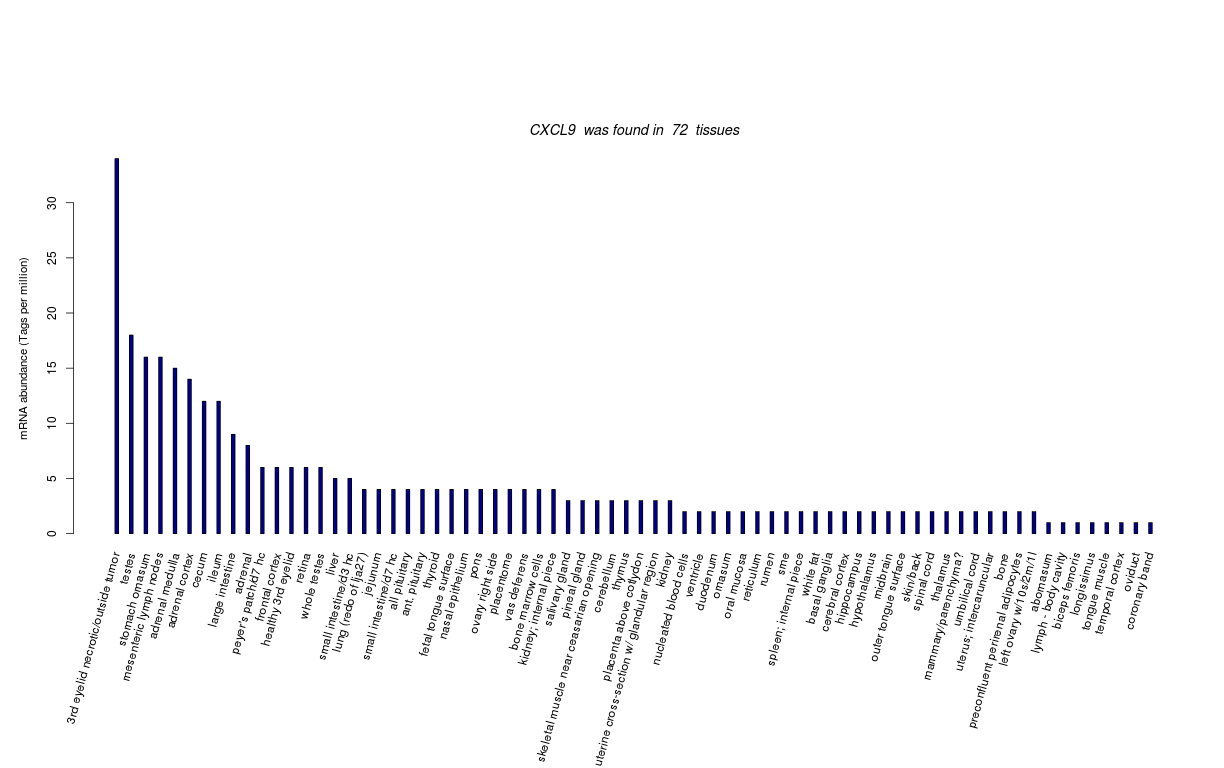
| Tag Count based mRNA-Abundances across 87 different Tissues (TPM).
Based on Data from Bovine Gene Atlas |
(Move your mouse over the image to view a more detailed version) |
||||||||||||||||||