| Bos taurus Gene: SUMF1 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Summary | |||||||||||
| InnateDB Gene | IDBG-644833.3 | ||||||||||
| Last Modified | 2014-10-13 [Report errors or provide feedback] | ||||||||||
| Gene Symbol | SUMF1 | ||||||||||
| Gene Name | Sulfatase-modifying factor 1 | ||||||||||
| Synonyms | |||||||||||
| Species | Bos taurus | ||||||||||
| Ensembl Gene | ENSBTAG00000039855 | ||||||||||
| Encoded Proteins |
Sulfatase-modifying factor 1
|
||||||||||
| Protein Structure |

|
||||||||||
| Useful resources | Stemformatics EHFPI ImmGen | ||||||||||
| Entrez Gene | |||||||||||
| Summary |
This gene does not have any Entrez summary - the following is the summary from its human ortholog ENSG00000144455:
This gene encodes an enzyme that catalyzes the hydrolysis of sulfate esters by oxidizing a cysteine residue in the substrate sulfatase to an active site 3-oxoalanine residue, which is also known as C-alpha-formylglycine. Mutations in this gene cause multiple sulfatase deficiency, a lysosomal storage disorder. Alternative splicing results in multiple transcript variants. [provided by RefSeq, Sep 2009] |
||||||||||
| Gene Information | |||||||||||
| Type | Protein coding | ||||||||||
| Genomic Location | Chromosome 22:21994945-22088653 | ||||||||||
| Strand | Forward strand | ||||||||||
| Band | |||||||||||
| Transcripts |
|
||||||||||
| Interactions | |||||||||||
| Number of Interactions |
This gene and/or its encoded proteins are associated with 0 experimentally validated interaction(s) in this database.
They are also associated with 1 interaction(s) predicted by orthology.
|
||||||||||
| Gene Ontology | |||||||||||
Molecular Function |
|
||||||||||
| Biological Process |
|
||||||||||
| Cellular Component |
|
||||||||||
| Orthologs | |||||||||||
|
Species
Homo sapiens
Mus musculus
|
Gene ID
Gene Order
|
||||||||||
| Pathways | |||||||||||
| NETPATH | |||||||||||
| REACTOME |
Glycosphingolipid metabolism pathway
The activation of arylsulfatases pathway
Metabolism of proteins pathway
Post-translational protein modification pathway
Sphingolipid metabolism pathway
Metabolism of lipids and lipoproteins pathway
Metabolism pathway
PTM: gamma carboxylation, hypusine formation and arylsulfatase activation pathway
|
||||||||||
| KEGG | |||||||||||
| INOH | |||||||||||
| PID NCI | |||||||||||
| Pathway Predictions based on Human Orthology Data | |||||||||||
| NETPATH | |||||||||||
| REACTOME |
The activation of arylsulfatases pathway
Glycosphingolipid metabolism pathway
Sphingolipid metabolism pathway
Metabolism of lipids and lipoproteins pathway
Post-translational protein modification pathway
PTM: gamma carboxylation, hypusine formation and arylsulfatase activation pathway
Metabolism of proteins pathway
Metabolism pathway
The activation of arylsulfatases pathway
Glycosphingolipid metabolism pathway
Metabolism pathway
Sphingolipid metabolism pathway
Metabolism of proteins pathway
Post-translational protein modification pathway
Metabolism of lipids and lipoproteins pathway
PTM: gamma carboxylation, hypusine formation and arylsulfatase activation pathway
|
||||||||||
| KEGG |
Lysosome pathway
Lysosome pathway
|
||||||||||
| INOH | |||||||||||
| PID NCI | |||||||||||
| Cross-References | |||||||||||
| SwissProt | Q0P5L5 | ||||||||||
| TrEMBL | |||||||||||
| UniProt Splice Variant | |||||||||||
| Entrez Gene | 536435 | ||||||||||
| UniGene | Bt.30055 Bt.88474 Bt.88475 | ||||||||||
| RefSeq | NM_001076076 | ||||||||||
| HUGO | |||||||||||
| OMIM | |||||||||||
| CCDS | |||||||||||
| HPRD | |||||||||||
| IMGT | |||||||||||
| EMBL | BC119885 | ||||||||||
| GenPept | AAI19886 | ||||||||||
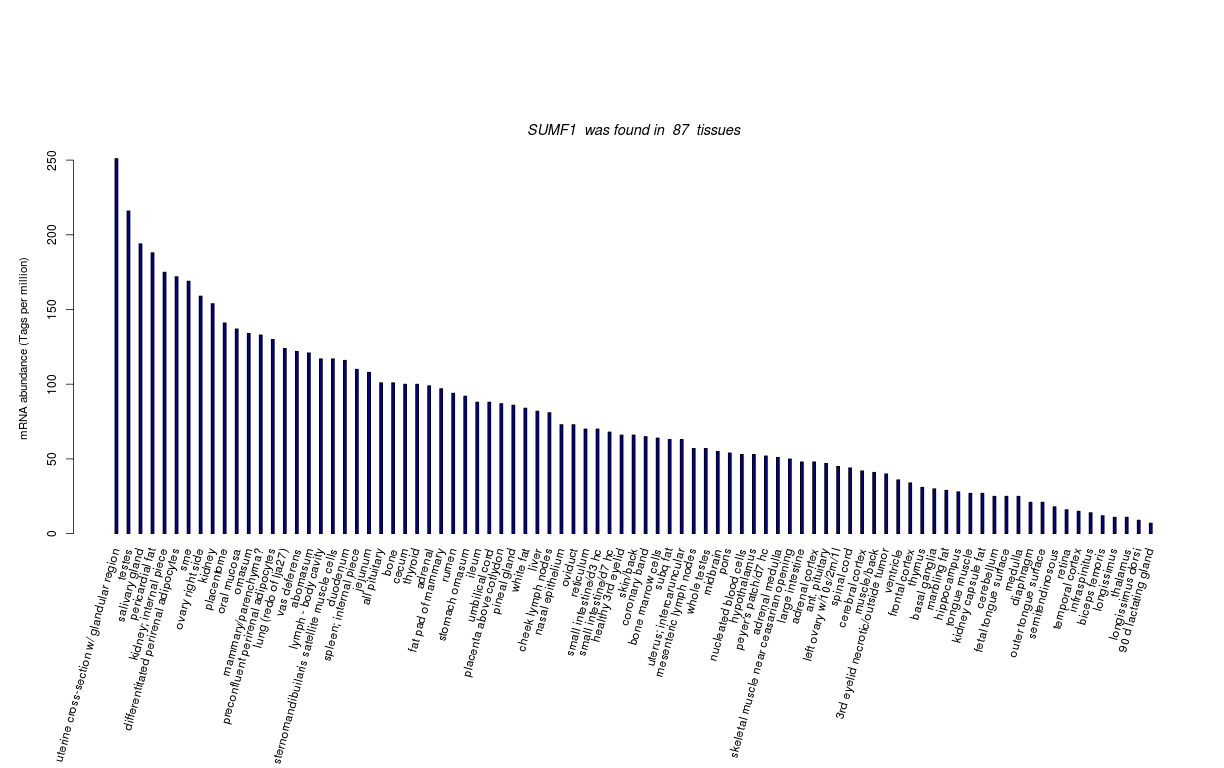
| RNA Seq Atlas | 536435 | ||||||||||
| Transcript Frequencies | |||||||||||
| Tag Count based mRNA-Abundances across 87 different Tissues (TPM).
Based on Data from Bovine Gene Atlas |
(Move your mouse over the image to view a more detailed version) |
||||||||||