| Bos taurus Gene: CD81 | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Summary | |||||||||||||||
| InnateDB Gene | IDBG-645214.3 | ||||||||||||||
| Last Modified | 2014-10-13 [Report errors or provide feedback] | ||||||||||||||
| Gene Symbol | CD81 | ||||||||||||||
| Gene Name | CD81 antigen | ||||||||||||||
| Synonyms | |||||||||||||||
| Species | Bos taurus | ||||||||||||||
| Ensembl Gene | ENSBTAG00000047495 | ||||||||||||||
| Encoded Proteins |
CD81 antigen
|
||||||||||||||
| Protein Structure |

|
||||||||||||||
| Useful resources | Stemformatics EHFPI ImmGen | ||||||||||||||
| InnateDB Annotation from Orthologs | |||||||||||||||
| Summary |
[Mus musculus] Cd81 inhibits Rac1/Stat1 activation and negatively regulates the defence mechanisms to Listeria monocytogenes infection.
|
||||||||||||||
| Entrez Gene | |||||||||||||||
| Summary |
This gene does not have any Entrez summary - the following is the summary from its human ortholog ENSG00000110651:
The protein encoded by this gene is a member of the transmembrane 4 superfamily, also known as the tetraspanin family. Most of these members are cell-surface proteins that are characterized by the presence of four hydrophobic domains. The proteins mediate signal transduction events that play a role in the regulation of cell development, activation, growth and motility. This encoded protein is a cell surface glycoprotein that is known to complex with integrins. This protein appears to promote muscle cell fusion and support myotube maintenance. Also it may be involved in signal transduction. This gene is localized in the tumor-suppressor gene region and thus it is a candidate gene for malignancies. [provided by RefSeq, Jul 2008] The protein encoded by this gene is a member of the transmembrane 4 superfamily, also known as the tetraspanin family. Most of these members are cell-surface proteins that are characterized by the presence of four hydrophobic domains. The proteins mediate signal transduction events that play a role in the regulation of cell development, activation, growth and motility. This encoded protein is a cell surface glycoprotein that is known to complex with integrins. This protein appears to promote muscle cell fusion and support myotube maintenance. Also it may be involved in signal transduction. This gene is localized in the tumor-suppressor gene region and thus it is a candidate gene for malignancies. Two transcript variants encoding different isoforms have been found for this gene. [provided by RefSeq, Jul 2014] |
||||||||||||||
| Gene Information | |||||||||||||||
| Type | Protein coding | ||||||||||||||
| Genomic Location | Chromosome 29:49842969-49848639 | ||||||||||||||
| Strand | Reverse strand | ||||||||||||||
| Band | |||||||||||||||
| Transcripts |
|
||||||||||||||
| Interactions | |||||||||||||||
| Number of Interactions |
This gene and/or its encoded proteins are associated with 0 experimentally validated interaction(s) in this database.
They are also associated with 22 interaction(s) predicted by orthology.
|
||||||||||||||
| Gene Ontology | |||||||||||||||
Molecular Function |
|
||||||||||||||
| Biological Process |
|
||||||||||||||
| Cellular Component |
|
||||||||||||||
| Orthologs | |||||||||||||||
|
Species
Homo sapiens
Mus musculus
|
Gene ID
Gene Order
|
||||||||||||||
| Pathways | |||||||||||||||
| NETPATH | |||||||||||||||
| REACTOME |
Immune System pathway
Adaptive Immune System pathway
Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell pathway
|
||||||||||||||
| KEGG | |||||||||||||||
| INOH | |||||||||||||||
| PID NCI | |||||||||||||||
| Pathway Predictions based on Human Orthology Data | |||||||||||||||
| NETPATH |
BCR pathway
|
||||||||||||||
| REACTOME |
Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell pathway
Adaptive Immune System pathway
Immune System pathway
Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell pathway
Immune System pathway
Adaptive Immune System pathway
|
||||||||||||||
| KEGG |
B cell receptor signaling pathway pathway
Malaria pathway
Hepatitis C pathway
B cell receptor signaling pathway pathway
Malaria pathway
Hepatitis C pathway
|
||||||||||||||
| INOH | |||||||||||||||
| PID NCI |
Beta1 integrin cell surface interactions
Alpha4 beta1 integrin signaling events
|
||||||||||||||
| Cross-References | |||||||||||||||
| SwissProt | Q3ZCD0 | ||||||||||||||
| TrEMBL | G3MYH4 | ||||||||||||||
| UniProt Splice Variant | |||||||||||||||
| Entrez Gene | 511435 | ||||||||||||||
| UniGene | Bt.51424 | ||||||||||||||
| RefSeq | NM_001035099 | ||||||||||||||
| HUGO | |||||||||||||||
| OMIM | |||||||||||||||
| CCDS | |||||||||||||||
| HPRD | |||||||||||||||
| IMGT | |||||||||||||||
| EMBL | BC102513 DAAA02063726 DAAA02063727 | ||||||||||||||
| GenPept | AAI02514 | ||||||||||||||
| RNA Seq Atlas | 511435 | ||||||||||||||
| Transcript Frequencies | |||||||||||||||
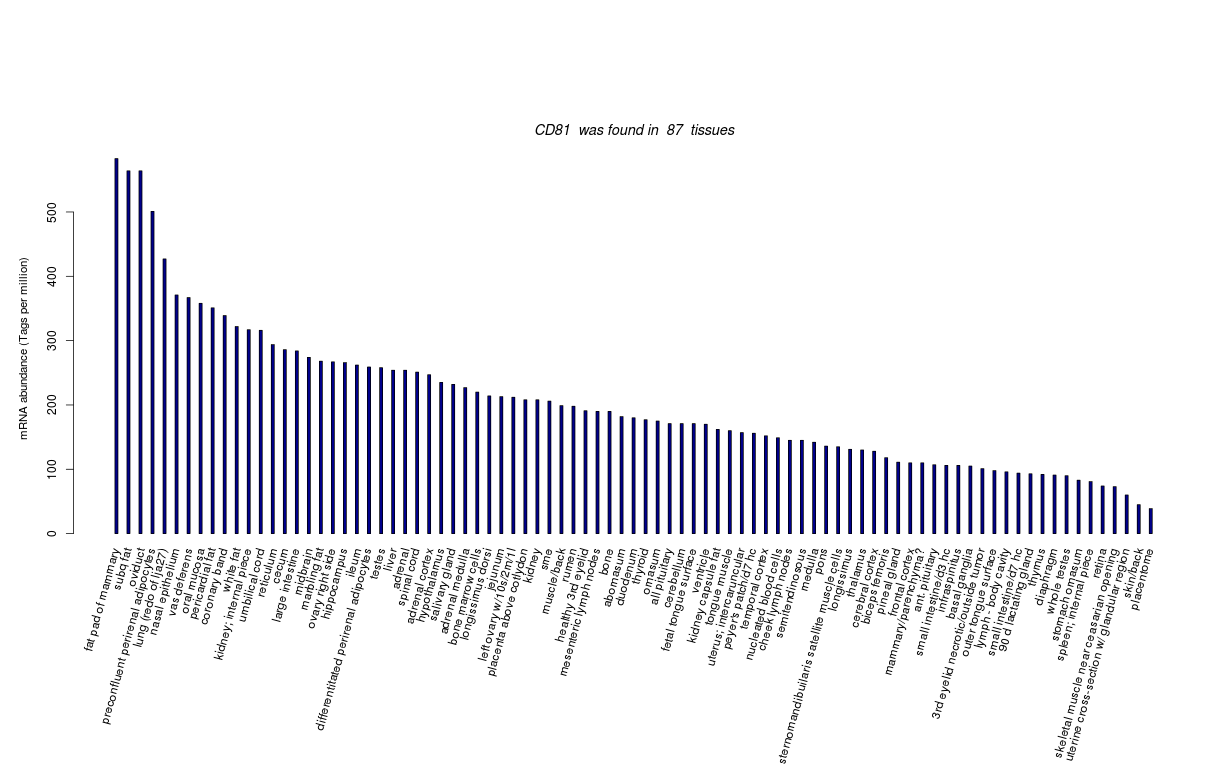
| Tag Count based mRNA-Abundances across 87 different Tissues (TPM).
Based on Data from Bovine Gene Atlas |
(Move your mouse over the image to view a more detailed version) |
||||||||||||||