| Bos taurus Gene: PTBP1 | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Summary | |||||||||||||||
| InnateDB Gene | IDBG-645533.3 | ||||||||||||||
| Last Modified | 2014-10-13 [Report errors or provide feedback] | ||||||||||||||
| Gene Symbol | PTBP1 | ||||||||||||||
| Gene Name | Polypyrimidine tract-binding protein 1 | ||||||||||||||
| Synonyms | |||||||||||||||
| Species | Bos taurus | ||||||||||||||
| Ensembl Gene | ENSBTAG00000045828 | ||||||||||||||
| Encoded Proteins |
Polypyrimidine tract-binding protein 1
Polypyrimidine tract-binding protein 1
|
||||||||||||||
| Protein Structure |

|
||||||||||||||
| Useful resources | Stemformatics EHFPI ImmGen | ||||||||||||||
| Entrez Gene | |||||||||||||||
| Summary |
This gene does not have any Entrez summary - the following is the summary from its human ortholog ENSG00000011304:
microRNAs (miRNAs) are short (20-24 nt) non-coding RNAs that are involved in post-transcriptional regulation of gene expression in multicellular organisms by affecting both the stability and translation of mRNAs. miRNAs are transcribed by RNA polymerase II as part of capped and polyadenylated primary transcripts (pri-miRNAs) that can be either protein-coding or non-coding. The primary transcript is cleaved by the Drosha ribonuclease III enzyme to produce an approximately 70-nt stem-loop precursor miRNA (pre-miRNA), which is further cleaved by the cytoplasmic Dicer ribonuclease to generate the mature miRNA and antisense miRNA star (miRNA*) products. The mature miRNA is incorporated into a RNA-induced silencing complex (RISC), which recognizes target mRNAs through imperfect base pairing with the miRNA and most commonly results in translational inhibition or destabilization of the target mRNA. The RefSeq represents the predicted microRNA stem-loop. [provided by RefSeq, Sep 2009] This gene belongs to the subfamily of ubiquitously expressed heterogeneous nuclear ribonucleoproteins (hnRNPs). The hnRNPs are RNA-binding proteins and they complex with heterogeneous nuclear RNA (hnRNA). These proteins are associated with pre-mRNAs in the nucleus and appear to influence pre-mRNA processing and other aspects of mRNA metabolism and transport. While all of the hnRNPs are present in the nucleus, some seem to shuttle between the nucleus and the cytoplasm. The hnRNP proteins have distinct nucleic acid binding properties. The protein encoded by this gene has four repeats of quasi-RNA recognition motif (RRM) domains that bind RNAs. This protein binds to the intronic polypyrimidine tracts that requires pre-mRNA splicing and acts via the protein degradation ubiquitin-proteasome pathway. It may also promote the binding of U2 snRNP to pre-mRNAs. This protein is localized in the nucleoplasm and it is also detected in the perinucleolar structure. Alternatively spliced transcript variants encoding different isoforms have been described. [provided by RefSeq, Jul 2008] This gene belongs to the subfamily of ubiquitously expressed heterogeneous nuclear ribonucleoproteins (hnRNPs). The hnRNPs are RNA-binding proteins and they complex with heterogeneous nuclear RNA (hnRNA). These proteins are associated with pre-mRNAs in the nucleus and appear to influence pre-mRNA processing and other aspects of mRNA metabolism and transport. While all of the hnRNPs are present in the nucleus, some seem to shuttle between the nucleus and the cytoplasm. The hnRNP proteins have distinct nucleic acid binding properties. The protein encoded by this gene has four repeats of quasi-RNA recognition motif (RRM) domains that bind RNAs. This protein binds to the intronic polypyrimidine tracts that requires pre-mRNA splicing and acts via the protein degradation ubiquitin-proteasome pathway. It may also promote the binding of U2 snRNP to pre-mRNAs. This protein is localized in the nucleoplasm and it is also detected in the perinucleolar structure. Alternatively spliced transcript variants encoding different isoforms have been described. [provided by RefSeq, Jul 2008] |
||||||||||||||
| Gene Information | |||||||||||||||
| Type | Protein coding | ||||||||||||||
| Genomic Location | Chromosome 7:45016275-45026949 | ||||||||||||||
| Strand | Forward strand | ||||||||||||||
| Band | |||||||||||||||
| Transcripts |
|
||||||||||||||
| Interactions | |||||||||||||||
| Number of Interactions |
This gene and/or its encoded proteins are associated with 0 experimentally validated interaction(s) in this database.
They are also associated with 58 interaction(s) predicted by orthology.
|
||||||||||||||
| Gene Ontology | |||||||||||||||
Molecular Function |
|
||||||||||||||
| Biological Process |
|
||||||||||||||
| Cellular Component |
|
||||||||||||||
| Orthologs | |||||||||||||||
|
Species
Homo sapiens
Mus musculus
|
Gene ID
Gene Order
|
||||||||||||||
| Pathways | |||||||||||||||
| NETPATH | |||||||||||||||
| REACTOME |
mRNA Splicing - Major Pathway pathway
mRNA Splicing pathway
Gene Expression pathway
Processing of Capped Intron-Containing Pre-mRNA pathway
|
||||||||||||||
| KEGG | |||||||||||||||
| INOH | |||||||||||||||
| PID NCI | |||||||||||||||
| Pathway Predictions based on Human Orthology Data | |||||||||||||||
| NETPATH | |||||||||||||||
| REACTOME |
mRNA Splicing - Major Pathway pathway
Processing of Capped Intron-Containing Pre-mRNA pathway
mRNA Splicing pathway
Gene Expression pathway
|
||||||||||||||
| KEGG | |||||||||||||||
| INOH | |||||||||||||||
| PID NCI | |||||||||||||||
| Cross-References | |||||||||||||||
| SwissProt | Q8WN55 | ||||||||||||||
| TrEMBL | A8YXY5 | ||||||||||||||
| UniProt Splice Variant | |||||||||||||||
| Entrez Gene | 282018 | ||||||||||||||
| UniGene | Bt.9031 | ||||||||||||||
| RefSeq | NM_174442 | ||||||||||||||
| HUGO | |||||||||||||||
| OMIM | |||||||||||||||
| CCDS | |||||||||||||||
| HPRD | |||||||||||||||
| IMGT | |||||||||||||||
| EMBL | AF445640 BC103383 DAAA02020232 | ||||||||||||||
| GenPept | AAI03384 AAL38169 | ||||||||||||||
| RNA Seq Atlas | 282018 | ||||||||||||||
| Transcript Frequencies | |||||||||||||||
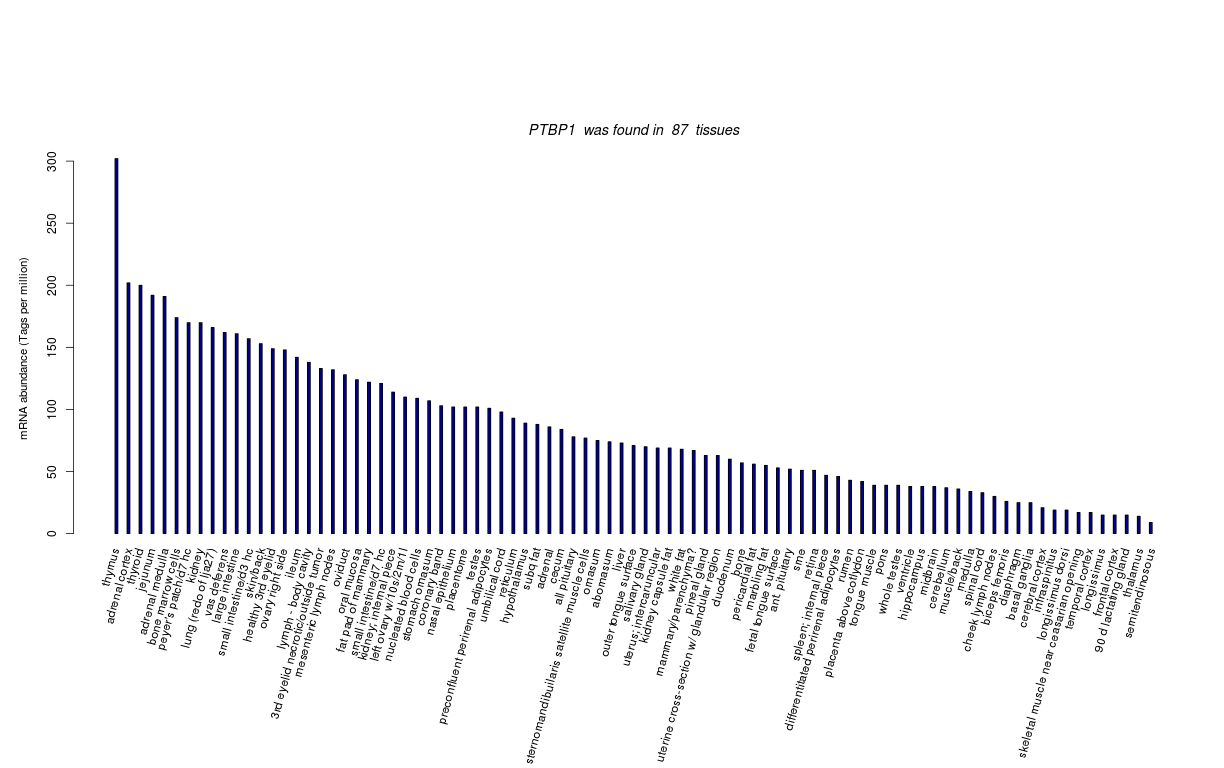
| Tag Count based mRNA-Abundances across 87 different Tissues (TPM).
Based on Data from Bovine Gene Atlas |
(Move your mouse over the image to view a more detailed version) |
||||||||||||||