| Bos taurus Gene: AUH | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Summary | |||||||||||||
| InnateDB Gene | IDBG-648347.3 | ||||||||||||
| Last Modified | 2014-10-13 [Report errors or provide feedback] | ||||||||||||
| Gene Symbol | AUH | ||||||||||||
| Gene Name | Uncharacterized protein | ||||||||||||
| Synonyms | |||||||||||||
| Species | Bos taurus | ||||||||||||
| Ensembl Gene | ENSBTAG00000044081 | ||||||||||||
| Encoded Proteins |
AU RNA binding protein/enoyl-CoA hydratase
|
||||||||||||
| Protein Structure | |||||||||||||
| Useful resources | Stemformatics EHFPI ImmGen | ||||||||||||
| Entrez Gene | |||||||||||||
| Summary |
This gene does not have any Entrez summary - the following is the summary from its human ortholog ENSG00000148090:
The methylglutaconyl-CoA hydratase, mitochondrial protein binds to the AU-rich element (ARE), a common element found in the 3' UTR of rapidly decaying mRNA such as c-fos, c-myc and granulocyte/ macrophage colony stimulating factor. ARE elements are involved in directing RNA to rapid degradation and deadenylation. AUH is also homologous to enol-CoA hydratase, an enzyme involved in fatty acid degradation, and has been shown to have intrinsic hydratase enzymatic activity. AUH is thus a bifunctional chimera between RNA binding and metabolic enzyme activity. A possible subcellular localization in the mitochondria has been demonstrated for the mouse homolog of this protein which shares 92% identity with the human protein. It has been suggested that AUH may have a novel role as a mitochondrial located AU-binding protein. Human AUH is expressed as a single mRNA species of 1.8 kb, and translated as a 40-kDa precursor protein which is subsequently processed to a 32-kDa mature form. [provided by RefSeq, May 2010] The methylglutaconyl-CoA hydratase, mitochondrial protein binds to the AU-rich element (ARE), a common element found in the 3\' UTR of rapidly decaying mRNA such as c-fos, c-myc and granulocyte/ macrophage colony stimulating factor. ARE elements are involved in directing RNA to rapid degradation and deadenylation. AUH is also homologous to enol-CoA hydratase, an enzyme involved in fatty acid degradation, and has been shown to have intrinsic hydratase enzymatic activity. AUH is thus a bifunctional chimera between RNA binding and metabolic enzyme activity. A possible subcellular localization in the mitochondria has been demonstrated for the mouse homolog of this protein which shares 92%% identity with the human protein. It has been suggested that AUH may have a novel role as a mitochondrial located AU-binding protein. Human AUH is expressed as a single mRNA species of 1.8 kb, and translated as a 40-kDa precursor protein which is subsequently processed to a 32-kDa mature form. [provided by RefSeq, May 2010] |
||||||||||||
| Gene Information | |||||||||||||
| Type | Protein coding | ||||||||||||
| Genomic Location | Chromosome 16:27058047-27058913 | ||||||||||||
| Strand | Forward strand | ||||||||||||
| Band | |||||||||||||
| Transcripts |
|
||||||||||||
| Interactions | |||||||||||||
| Number of Interactions |
This gene and/or its encoded proteins are associated with 0 experimentally validated interaction(s) in this database.
|
||||||||||||
| Gene Ontology | |||||||||||||
Molecular Function |
|
||||||||||||
| Biological Process |
|
||||||||||||
| Cellular Component |
|
||||||||||||
| Orthologs | |||||||||||||
|
Species
Homo sapiens
Mus musculus
|
Gene ID
Gene Order
|
||||||||||||
| Pathway Predictions based on Human Orthology Data | |||||||||||||
| NETPATH | |||||||||||||
| REACTOME |
Branched-chain amino acid catabolism pathway
Metabolism of amino acids and derivatives pathway
Metabolism pathway
Metabolism pathway
Metabolism of amino acids and derivatives pathway
Branched-chain amino acid catabolism pathway
|
||||||||||||
| KEGG |
Valine, leucine and isoleucine degradation pathway
Valine, leucine and isoleucine degradation pathway
|
||||||||||||
| INOH |
Valine Leucine Isoleucine degradation pathway
|
||||||||||||
| PID NCI | |||||||||||||
| Cross-References | |||||||||||||
| SwissProt | |||||||||||||
| TrEMBL | F1MGJ7 | ||||||||||||
| UniProt Splice Variant | |||||||||||||
| Entrez Gene | |||||||||||||
| UniGene | |||||||||||||
| RefSeq | |||||||||||||
| HUGO | HGNC:890 | ||||||||||||
| OMIM | |||||||||||||
| CCDS | |||||||||||||
| HPRD | |||||||||||||
| IMGT | |||||||||||||
| EMBL | DAAA02042622 | ||||||||||||
| GenPept | |||||||||||||
| RNA Seq Atlas | |||||||||||||
| Transcript Frequencies | |||||||||||||
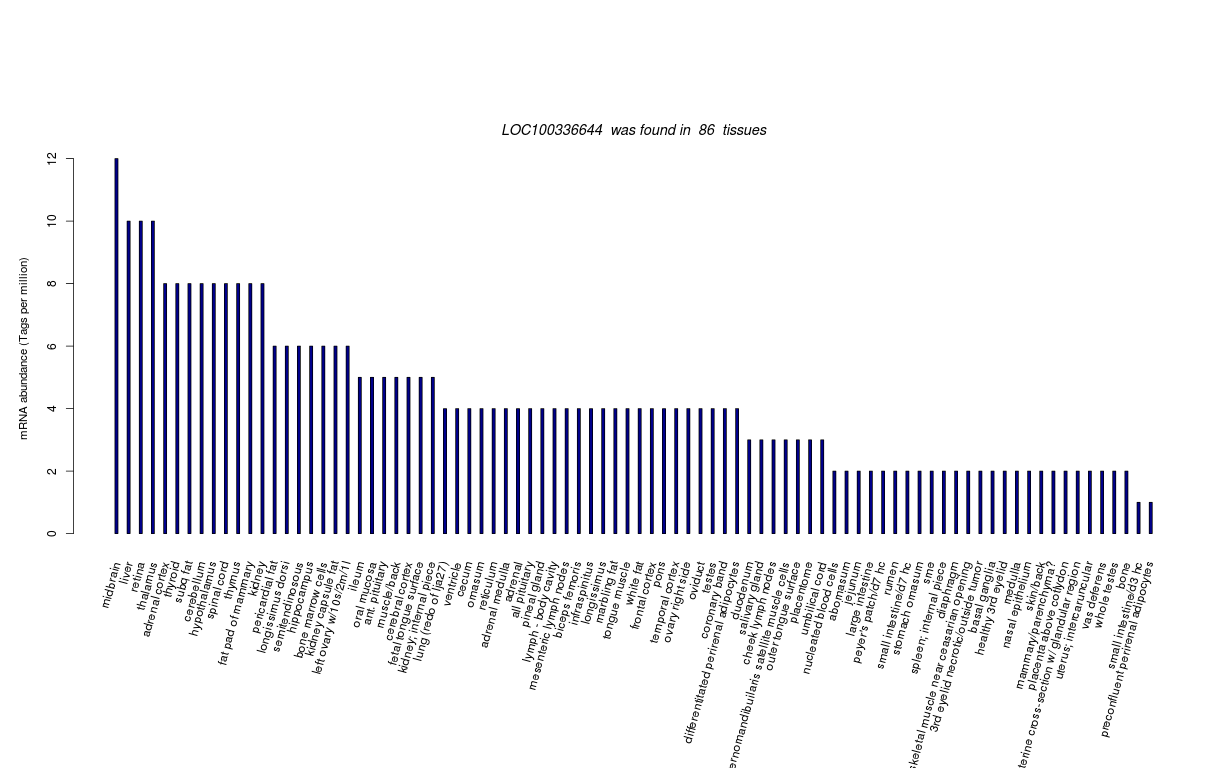
| Tag Count based mRNA-Abundances across 87 different Tissues (TPM).
Based on Data from Bovine Gene Atlas |
(Move your mouse over the image to view a more detailed version) |
||||||||||||