| Bos taurus Gene: BT.96766 | |||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Summary | |||||||||||||||||||||||||||||||||||||||||||||
| InnateDB Gene | IDBG-629054.3 | ||||||||||||||||||||||||||||||||||||||||||||
| Last Modified | 2014-10-13 [Report errors or provide feedback] | ||||||||||||||||||||||||||||||||||||||||||||
| Gene Symbol | BT.96766 | ||||||||||||||||||||||||||||||||||||||||||||
| Gene Name | Uncharacterized protein | ||||||||||||||||||||||||||||||||||||||||||||
| Synonyms | |||||||||||||||||||||||||||||||||||||||||||||
| Species | Bos taurus | ||||||||||||||||||||||||||||||||||||||||||||
| Ensembl Gene | ENSBTAG00000046409 | ||||||||||||||||||||||||||||||||||||||||||||
| Encoded Proteins |
Early growth response 2 protein
|
||||||||||||||||||||||||||||||||||||||||||||
| Protein Structure | |||||||||||||||||||||||||||||||||||||||||||||
| Useful resources | Stemformatics EHFPI ImmGen | ||||||||||||||||||||||||||||||||||||||||||||
| Entrez Gene | |||||||||||||||||||||||||||||||||||||||||||||
| Summary |
This gene does not have any Entrez summary - the following is the summary from its human ortholog ENSG00000122877:
The protein encoded by this gene is a transcription factor with three tandem C2H2-type zinc fingers. Defects in this gene are associated with Charcot-Marie-Tooth disease type 1D (CMT1D), Charcot-Marie-Tooth disease type 4E (CMT4E), and with Dejerine-Sottas syndrome (DSS). Multiple transcript variants encoding two different isoforms have been found for this gene. [provided by RefSeq, Oct 2008] |
||||||||||||||||||||||||||||||||||||||||||||
| Gene Information | |||||||||||||||||||||||||||||||||||||||||||||
| Type | Protein coding | ||||||||||||||||||||||||||||||||||||||||||||
| Genomic Location | Chromosome 28:19012870-19015927 | ||||||||||||||||||||||||||||||||||||||||||||
| Strand | Reverse strand | ||||||||||||||||||||||||||||||||||||||||||||
| Band | |||||||||||||||||||||||||||||||||||||||||||||
| Transcripts |
|
||||||||||||||||||||||||||||||||||||||||||||
| Interactions | |||||||||||||||||||||||||||||||||||||||||||||
| Number of Interactions |
This gene and/or its encoded proteins are associated with 0 experimentally validated interaction(s) in this database.
They are also associated with 23 interaction(s) predicted by orthology.
|
||||||||||||||||||||||||||||||||||||||||||||
| Gene Ontology | |||||||||||||||||||||||||||||||||||||||||||||
Molecular Function |
|
||||||||||||||||||||||||||||||||||||||||||||
| Biological Process |
|
||||||||||||||||||||||||||||||||||||||||||||
| Cellular Component |
|
||||||||||||||||||||||||||||||||||||||||||||
| Orthologs | |||||||||||||||||||||||||||||||||||||||||||||
|
Species
Homo sapiens
Mus musculus
|
Gene ID
Gene Order
|
||||||||||||||||||||||||||||||||||||||||||||
| Pathway Predictions based on Human Orthology Data | |||||||||||||||||||||||||||||||||||||||||||||
| NETPATH | |||||||||||||||||||||||||||||||||||||||||||||
| REACTOME |
Transcriptional regulation of white adipocyte differentiation pathway
Developmental Biology pathway
Transcriptional Regulation of Adipocyte Differentiation in 3T3-L1 Pre-adipocytes pathway
Transcriptional regulation of white adipocyte differentiation pathway
Mus musculus biological processes pathway
Developmental Biology pathway
|
||||||||||||||||||||||||||||||||||||||||||||
| KEGG | |||||||||||||||||||||||||||||||||||||||||||||
| INOH | |||||||||||||||||||||||||||||||||||||||||||||
| PID NCI |
Calcineurin-regulated NFAT-dependent transcription in lymphocytes
IL4-mediated signaling events
Validated transcriptional targets of TAp63 isoforms
|
||||||||||||||||||||||||||||||||||||||||||||
| Cross-References | |||||||||||||||||||||||||||||||||||||||||||||
| SwissProt | |||||||||||||||||||||||||||||||||||||||||||||
| TrEMBL | A5D9C8 G3N0C9 | ||||||||||||||||||||||||||||||||||||||||||||
| UniProt Splice Variant | |||||||||||||||||||||||||||||||||||||||||||||
| Entrez Gene | 100297981 | ||||||||||||||||||||||||||||||||||||||||||||
| UniGene | Bt.96766 | ||||||||||||||||||||||||||||||||||||||||||||
| RefSeq | |||||||||||||||||||||||||||||||||||||||||||||
| HUGO | |||||||||||||||||||||||||||||||||||||||||||||
| OMIM | |||||||||||||||||||||||||||||||||||||||||||||
| CCDS | |||||||||||||||||||||||||||||||||||||||||||||
| HPRD | |||||||||||||||||||||||||||||||||||||||||||||
| IMGT | |||||||||||||||||||||||||||||||||||||||||||||
| EMBL | BT030547 DAAA02061665 | ||||||||||||||||||||||||||||||||||||||||||||
| GenPept | ABQ12987 | ||||||||||||||||||||||||||||||||||||||||||||
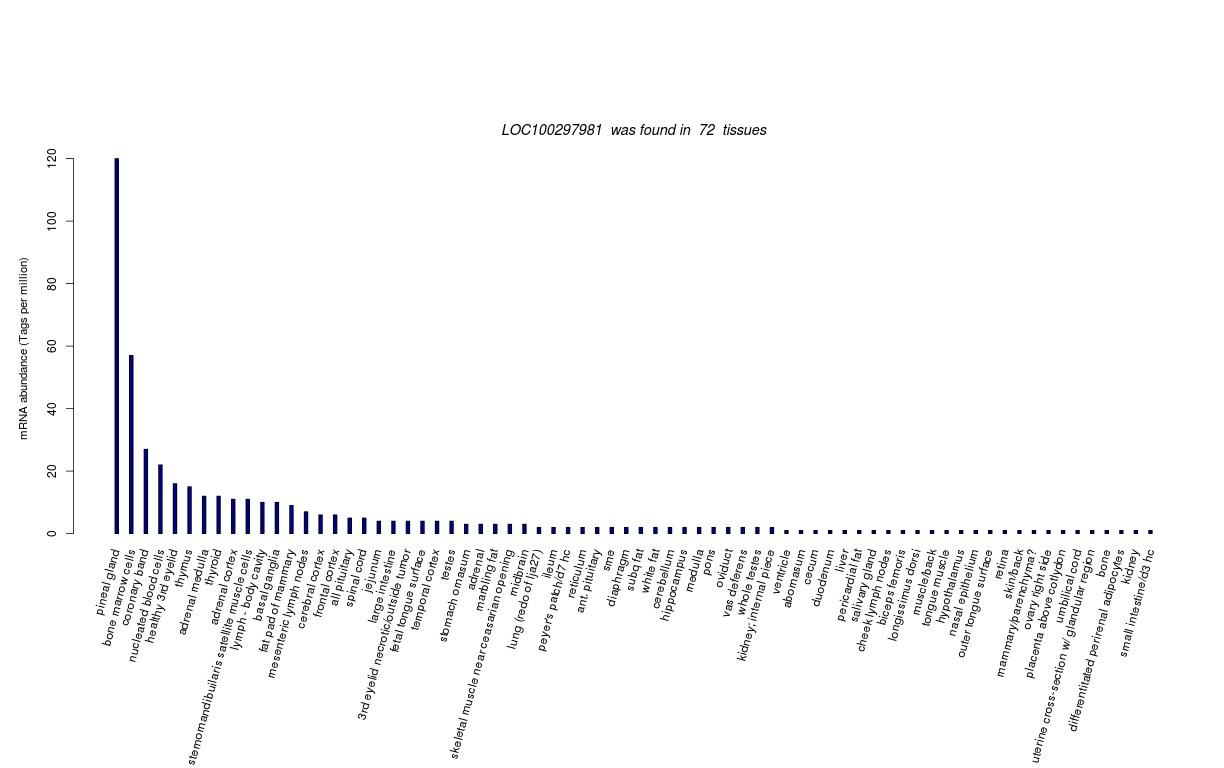
| RNA Seq Atlas | 100297981 | ||||||||||||||||||||||||||||||||||||||||||||
| Transcript Frequencies | |||||||||||||||||||||||||||||||||||||||||||||
| Tag Count based mRNA-Abundances across 87 different Tissues (TPM).
Based on Data from Bovine Gene Atlas |
(Move your mouse over the image to view a more detailed version) |
||||||||||||||||||||||||||||||||||||||||||||