| Bos taurus Gene: CCL2 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Summary | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| InnateDB Gene | IDBG-630954.3 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Last Modified | 2014-10-13 [Report errors or provide feedback] | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Gene Symbol | CCL2 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Gene Name | C-C motif chemokine 2 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Synonyms | MCP-1; MCP-1A; MCP1; MCP1A; SCYA2 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Species | Bos taurus | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Ensembl Gene | ENSBTAG00000037811 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Encoded Proteins |
C-C motif chemokine 2
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Protein Structure |

|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Useful resources | Stemformatics EHFPI ImmGen | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Entrez Gene | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Summary | Currently no Entrez Summary Available. You might want to check the Summary Sections of the Orthologs. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Gene Information | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Type | Protein coding | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Genomic Location | Chromosome 19:16232968-16234839 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Strand | Reverse strand | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Band | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Transcripts |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Interactions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Number of Interactions |
This gene and/or its encoded proteins are associated with 0 experimentally validated interaction(s) in this database.
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Gene Ontology | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Molecular Function |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Biological Process |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Cellular Component |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Orthologs | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| No orthologs found for this gene | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Pathways | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| NETPATH | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| REACTOME |
PERK regulates gene expression pathway
Peptide ligand-binding receptors pathway
Signaling by GPCR pathway
ATF4 activates genes pathway
Signal Transduction pathway
Metabolism of proteins pathway
GPCR ligand binding pathway
Class A/1 (Rhodopsin-like receptors) pathway
Unfolded Protein Response (UPR) pathway
Chemokine receptors bind chemokines pathway
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| KEGG | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| INOH | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| PID NCI | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Cross-References | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| SwissProt | P28291 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| TrEMBL | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| UniProt Splice Variant | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Entrez Gene | 281043 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| UniGene | Bt.2408 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| RefSeq | NM_174006 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| HUGO | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| OMIM | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| CCDS | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| HPRD | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| IMGT | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| EMBL | EU276059 L32659 M84602 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| GenPept | AAA30651 AAA60956 ABX72057 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| RNA Seq Atlas | 281043 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Transcript Frequencies | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
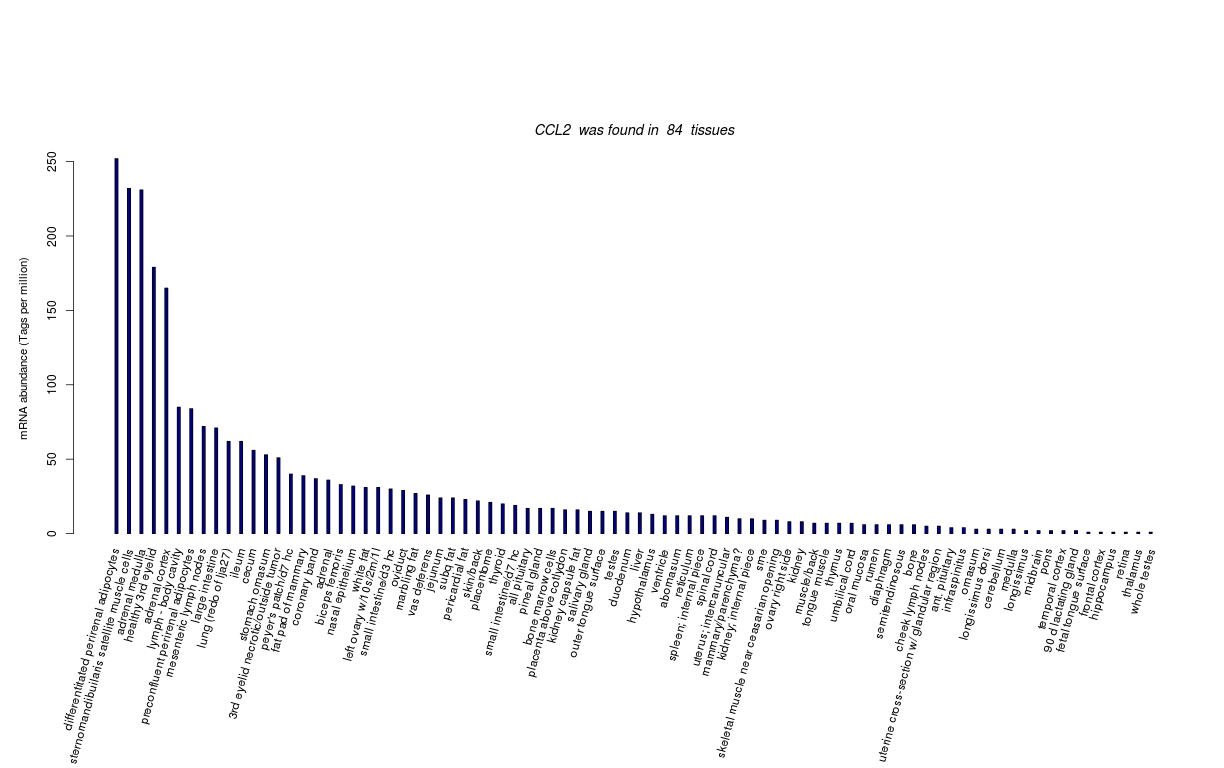
| Tag Count based mRNA-Abundances across 87 different Tissues (TPM).
Based on Data from Bovine Gene Atlas |
(Move your mouse over the image to view a more detailed version) |
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||