| Bos taurus Gene: ATRX | |||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Summary | |||||||||||||||||||||||||||||||||
| InnateDB Gene | IDBG-635337.3 | ||||||||||||||||||||||||||||||||
| Last Modified | 2014-10-13 [Report errors or provide feedback] | ||||||||||||||||||||||||||||||||
| Gene Symbol | ATRX | ||||||||||||||||||||||||||||||||
| Gene Name | Uncharacterized protein | ||||||||||||||||||||||||||||||||
| Synonyms | |||||||||||||||||||||||||||||||||
| Species | Bos taurus | ||||||||||||||||||||||||||||||||
| Ensembl Gene | ENSBTAG00000038434 | ||||||||||||||||||||||||||||||||
| Encoded Proteins |
alpha thalassemia/mental retardation syndrome X-linked
|
||||||||||||||||||||||||||||||||
| Protein Structure | |||||||||||||||||||||||||||||||||
| Useful resources | Stemformatics EHFPI ImmGen | ||||||||||||||||||||||||||||||||
| Entrez Gene | |||||||||||||||||||||||||||||||||
| Summary |
This gene does not have any Entrez summary - the following is the summary from its human ortholog ENSG00000085224:
The protein encoded by this gene contains an ATPase/helicase domain, and thus it belongs to the SWI/SNF family of chromatin remodeling proteins. The mutations of this gene are associated with an X-linked mental retardation (XLMR) syndrome most often accompanied by alpha-thalassemia (ATRX) syndrome. These mutations have been shown to cause diverse changes in the pattern of DNA methylation, which may provide a link between chromatin remodeling, DNA methylation, and gene expression in developmental processes. This protein is found to undergo cell cycle-dependent phosphorylation, which regulates its nuclear matrix and chromatin association, and suggests its involvement in the gene regulation at interphase and chromosomal segregation in mitosis. Multiple alternatively spliced transcript variants encoding distinct isoforms have been reported. [provided by RefSeq, Jul 2008] The protein encoded by this gene contains an ATPase/helicase domain, and thus it belongs to the SWI/SNF family of chromatin remodeling proteins. This protein is found to undergo cell cycle-dependent phosphorylation, which regulates its nuclear matrix and chromatin association, and suggests its involvement in the gene regulation at interphase and chromosomal segregation in mitosis. Mutations in this gene are associated with an X-linked mental retardation (XLMR) syndrome most often accompanied by alpha-thalassemia (ATRX) syndrome. These mutations have been shown to cause diverse changes in the pattern of DNA methylation, which may provide a link between chromatin remodeling, DNA methylation, and gene expression in developmental processes. Multiple alternatively spliced transcript variants encoding distinct isoforms have been reported. [provided by RefSeq, Aug 2013] |
||||||||||||||||||||||||||||||||
| Gene Information | |||||||||||||||||||||||||||||||||
| Type | Protein coding | ||||||||||||||||||||||||||||||||
| Genomic Location | Chromosome X:79634445-79917815 | ||||||||||||||||||||||||||||||||
| Strand | Forward strand | ||||||||||||||||||||||||||||||||
| Band | |||||||||||||||||||||||||||||||||
| Transcripts |
|
||||||||||||||||||||||||||||||||
| Interactions | |||||||||||||||||||||||||||||||||
| Number of Interactions |
This gene and/or its encoded proteins are associated with 0 experimentally validated interaction(s) in this database.
They are also associated with 21 interaction(s) predicted by orthology.
|
||||||||||||||||||||||||||||||||
| Gene Ontology | |||||||||||||||||||||||||||||||||
Molecular Function |
|
||||||||||||||||||||||||||||||||
| Biological Process |
|
||||||||||||||||||||||||||||||||
| Cellular Component |
|
||||||||||||||||||||||||||||||||
| Orthologs | |||||||||||||||||||||||||||||||||
|
Species
Homo sapiens
Mus musculus
|
Gene ID
Gene Order
|
||||||||||||||||||||||||||||||||
| Pathway Predictions based on Human Orthology Data | |||||||||||||||||||||||||||||||||
| NETPATH |
TSLP pathway
|
||||||||||||||||||||||||||||||||
| REACTOME | |||||||||||||||||||||||||||||||||
| KEGG | |||||||||||||||||||||||||||||||||
| INOH | |||||||||||||||||||||||||||||||||
| PID NCI | |||||||||||||||||||||||||||||||||
| Cross-References | |||||||||||||||||||||||||||||||||
| SwissProt | |||||||||||||||||||||||||||||||||
| TrEMBL | F1MQ85 | ||||||||||||||||||||||||||||||||
| UniProt Splice Variant | |||||||||||||||||||||||||||||||||
| Entrez Gene | 514473 | ||||||||||||||||||||||||||||||||
| UniGene | |||||||||||||||||||||||||||||||||
| RefSeq | XM_002699982 XM_002699983 XM_002707277 XM_005200750 XM_005200751 XM_005200752 XM_005200753 XM_005200754 XM_005200755 XM_005227973 XM_005227974 XM_005227975 XM_005227976 XM_005227977 XM_005227978 XM_592333 | ||||||||||||||||||||||||||||||||
| HUGO | HGNC:886 | ||||||||||||||||||||||||||||||||
| OMIM | |||||||||||||||||||||||||||||||||
| CCDS | |||||||||||||||||||||||||||||||||
| HPRD | |||||||||||||||||||||||||||||||||
| IMGT | |||||||||||||||||||||||||||||||||
| EMBL | DAAA02072500 DAAA02072501 DAAA02072502 DAAA02072503 DAAA02072504 DAAA02072505 DAAA02072506 DAAA02072507 | ||||||||||||||||||||||||||||||||
| GenPept | |||||||||||||||||||||||||||||||||
| RNA Seq Atlas | 514473 | ||||||||||||||||||||||||||||||||
| Transcript Frequencies | |||||||||||||||||||||||||||||||||
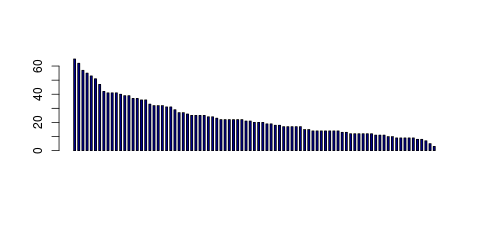
| Tag Count based mRNA-Abundances across 87 different Tissues (TPM).
Based on Data from Bovine Gene Atlas |
(Move your mouse over the image to view a more detailed version) |
||||||||||||||||||||||||||||||||