| Bos taurus Gene: GSTT1 | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Summary | |||||||||
| InnateDB Gene | IDBG-637112.3 | ||||||||
| Last Modified | 2014-10-13 [Report errors or provide feedback] | ||||||||
| Gene Symbol | GSTT1 | ||||||||
| Gene Name | Glutathione S-transferase theta-1 | ||||||||
| Synonyms | |||||||||
| Species | Bos taurus | ||||||||
| Ensembl Gene | ENSBTAG00000040298 | ||||||||
| Encoded Proteins |
Glutathione S-transferase theta-1
|
||||||||
| Protein Structure |

|
||||||||
| Useful resources | Stemformatics EHFPI ImmGen | ||||||||
| Entrez Gene | |||||||||
| Summary |
This gene does not have any Entrez summary - the following is the summary from its human ortholog null:
Glutathione S-transferase (GST) theta 1 (GSTT1) is a member of a superfamily of proteins that catalyze the conjugation of reduced glutathione to a variety of electrophilic and hydrophobic compounds. Human GSTs can be divided into five main classes: alpha, mu, pi, theta, and zeta. The theta class includes GSTT1 and GSTT2. The GSTT1 and GSTT2 share 55% amino acid sequence identity and both of them were claimed to have an important role in human carcinogenesis. The GSTT1 gene is located approximately 50kb away from the GSTT2 gene. The GSTT1 and GSTT2 genes have a similar structure, being composed of five exons with identical exon/intron boundaries. [provided by RefSeq, Jul 2008] |
||||||||
| Gene Information | |||||||||
| Type | Protein coding | ||||||||
| Genomic Location | Chromosome 17:73300145-73307793 | ||||||||
| Strand | Forward strand | ||||||||
| Band | |||||||||
| Transcripts |
|
||||||||
| Interactions | |||||||||
| Number of Interactions |
This gene and/or its encoded proteins are associated with 0 experimentally validated interaction(s) in this database.
|
||||||||
| Gene Ontology | |||||||||
Molecular Function |
|
||||||||
| Biological Process |
|
||||||||
| Cellular Component |
|
||||||||
| Orthologs | |||||||||
|
Species
Homo sapiens
Mus musculus
|
Gene ID
Gene Order
|
||||||||
| Pathways | |||||||||
| NETPATH | |||||||||
| REACTOME |
Glutathione conjugation pathway
Biological oxidations pathway
Metabolism pathway
Phase II conjugation pathway
|
||||||||
| KEGG | |||||||||
| INOH | |||||||||
| PID NCI | |||||||||
| Pathway Predictions based on Human Orthology Data | |||||||||
| NETPATH | |||||||||
| REACTOME |
Phase II conjugation pathway
Metabolism pathway
Glutathione conjugation pathway
Biological oxidations pathway
|
||||||||
| KEGG |
Glutathione metabolism pathway
Metabolism of xenobiotics by cytochrome P450 pathway
Drug metabolism pathway
|
||||||||
| INOH | |||||||||
| PID NCI | |||||||||
| Cross-References | |||||||||
| SwissProt | Q2NL00 | ||||||||
| TrEMBL | |||||||||
| UniProt Splice Variant | |||||||||
| Entrez Gene | 517724 | ||||||||
| UniGene | Bt.23204 | ||||||||
| RefSeq | NM_001046232 | ||||||||
| HUGO | |||||||||
| OMIM | |||||||||
| CCDS | |||||||||
| HPRD | |||||||||
| IMGT | |||||||||
| EMBL | BC111289 | ||||||||
| GenPept | AAI11290 | ||||||||
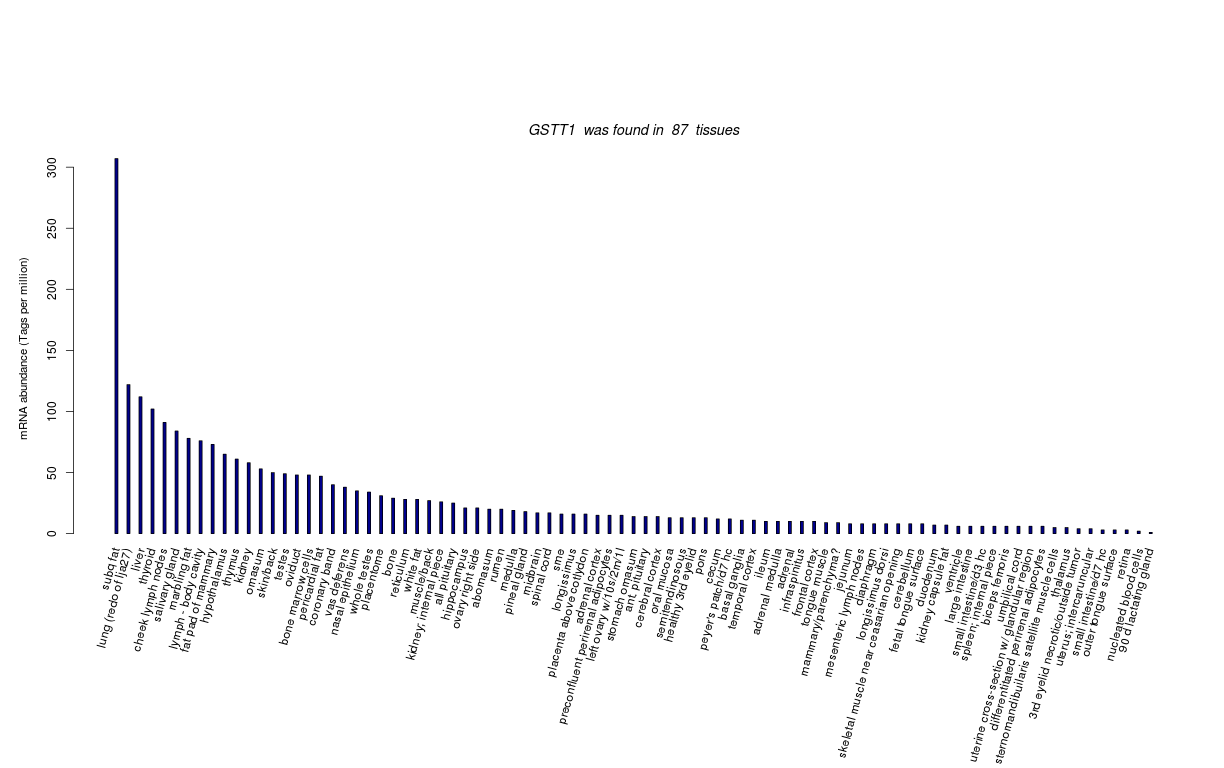
| RNA Seq Atlas | 517724 | ||||||||
| Transcript Frequencies | |||||||||
| Tag Count based mRNA-Abundances across 87 different Tissues (TPM).
Based on Data from Bovine Gene Atlas |
(Move your mouse over the image to view a more detailed version) |
||||||||