| Bos taurus Gene: NLRP12 | |||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Summary | |||||||||||||||||||||||||||||||||||
| InnateDB Gene | IDBG-641617.3 | ||||||||||||||||||||||||||||||||||
| Last Modified | 2014-10-13 [Report errors or provide feedback] | ||||||||||||||||||||||||||||||||||
| Gene Symbol | NLRP12 | ||||||||||||||||||||||||||||||||||
| Gene Name | NACHT, LRR and PYD domains-containing protein 12 | ||||||||||||||||||||||||||||||||||
| Synonyms | |||||||||||||||||||||||||||||||||||
| Species | Bos taurus | ||||||||||||||||||||||||||||||||||
| Ensembl Gene | ENSBTAG00000038149 | ||||||||||||||||||||||||||||||||||
| Encoded Proteins |
NLR family, pyrin domain containing 12
NLR family, pyrin domain containing 12
|
||||||||||||||||||||||||||||||||||
| Protein Structure | |||||||||||||||||||||||||||||||||||
| Useful resources | Stemformatics EHFPI ImmGen | ||||||||||||||||||||||||||||||||||
| InnateDB Annotation from Orthologs | |||||||||||||||||||||||||||||||||||
| Summary |
[Homo sapiens] NLRP12 is a negative regulator of the NF-κB response in monocytes.
[Homo sapiens] NLRP12 negatively regulates non-canonical NF-κB pathway by inducing NIK degradation.
[Homo sapiens] NLRP12 is an antagonist of toll-like receptor-, tumour necrosis factor alpha-, and Mycobacterium tuberculosis-induced pro-inflammatory signals.
[Mus musculus] Nlrp12 is an intrinsic negative regulator of T-cell-mediated immunity. Altered NF-kB regulation and Il4 production are key mediators of Nlrp12-associated disease.
|
||||||||||||||||||||||||||||||||||
| Entrez Gene | |||||||||||||||||||||||||||||||||||
| Summary |
This gene does not have any Entrez summary - the following is the summary from its human ortholog ENSG00000142405:
This gene encodes a member of the CATERPILLER family of cytoplasmic proteins. The encoded protein, which contains an N-terminal pyrin domain, a NACHT domain, a NACHT-associated domain, and a C-terminus leucine-rich repeat region, functions as an attenuating factor of inflammation by suppressing inflammatory responses in activated monocytes. Mutations in this gene cause familial cold autoinflammatory syndrome type 2. Alternative splicing results in multiple transcript variants. [provided by RefSeq, Mar 2013] |
||||||||||||||||||||||||||||||||||
| Gene Information | |||||||||||||||||||||||||||||||||||
| Type | Protein coding | ||||||||||||||||||||||||||||||||||
| Genomic Location | Chromosome 18:61091348-61143063 | ||||||||||||||||||||||||||||||||||
| Strand | Forward strand | ||||||||||||||||||||||||||||||||||
| Band | |||||||||||||||||||||||||||||||||||
| Transcripts |
|
||||||||||||||||||||||||||||||||||
| Interactions | |||||||||||||||||||||||||||||||||||
| Number of Interactions |
This gene and/or its encoded proteins are associated with 0 experimentally validated interaction(s) in this database.
They are also associated with 8 interaction(s) predicted by orthology.
|
||||||||||||||||||||||||||||||||||
| Gene Ontology | |||||||||||||||||||||||||||||||||||
Molecular Function |
|
||||||||||||||||||||||||||||||||||
| Biological Process |
|
||||||||||||||||||||||||||||||||||
| Cellular Component |
|
||||||||||||||||||||||||||||||||||
| Orthologs | |||||||||||||||||||||||||||||||||||
|
Species
Homo sapiens
Mus musculus
|
Gene ID
Gene Order
|
||||||||||||||||||||||||||||||||||
| Cross-References | |||||||||||||||||||||||||||||||||||
| SwissProt | |||||||||||||||||||||||||||||||||||
| TrEMBL | |||||||||||||||||||||||||||||||||||
| UniProt Splice Variant | |||||||||||||||||||||||||||||||||||
| Entrez Gene | |||||||||||||||||||||||||||||||||||
| UniGene | Bt.101481 Bt.106406 | ||||||||||||||||||||||||||||||||||
| RefSeq | NM_001192551 | ||||||||||||||||||||||||||||||||||
| HUGO | |||||||||||||||||||||||||||||||||||
| OMIM | |||||||||||||||||||||||||||||||||||
| CCDS | |||||||||||||||||||||||||||||||||||
| HPRD | |||||||||||||||||||||||||||||||||||
| IMGT | |||||||||||||||||||||||||||||||||||
| EMBL | |||||||||||||||||||||||||||||||||||
| GenPept | |||||||||||||||||||||||||||||||||||
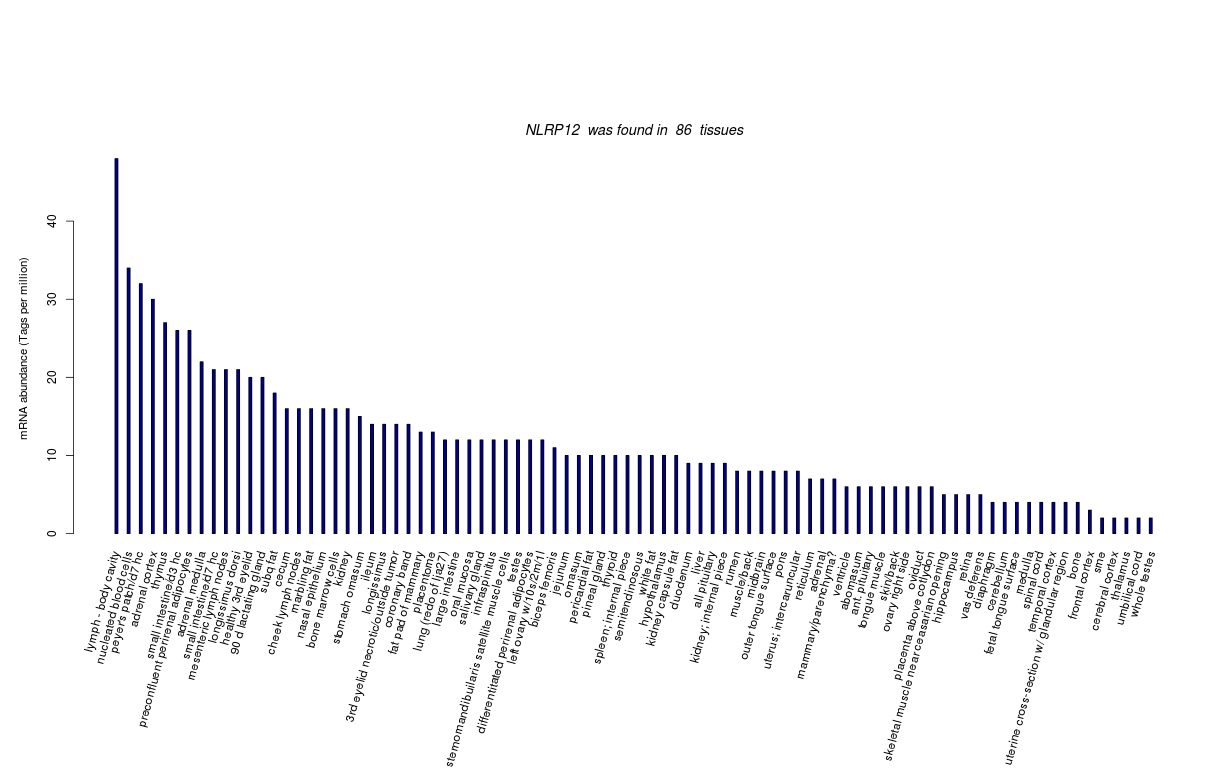
| RNA Seq Atlas | |||||||||||||||||||||||||||||||||||
| Transcript Frequencies | |||||||||||||||||||||||||||||||||||
| Tag Count based mRNA-Abundances across 87 different Tissues (TPM).
Based on Data from Bovine Gene Atlas |
(Move your mouse over the image to view a more detailed version) |
||||||||||||||||||||||||||||||||||