| Bos taurus Gene: ARHGEF18 | |||||||
|---|---|---|---|---|---|---|---|
| Summary | |||||||
| InnateDB Gene | IDBG-643626.3 | ||||||
| Last Modified | 2014-10-13 [Report errors or provide feedback] | ||||||
| Gene Symbol | ARHGEF18 | ||||||
| Gene Name | Uncharacterized protein | ||||||
| Synonyms | |||||||
| Species | Bos taurus | ||||||
| Ensembl Gene | ENSBTAG00000040507 | ||||||
| Encoded Proteins |
Rho/Rac guanine nucleotide exchange factor (GEF) 18
Rho/Rac guanine nucleotide exchange factor (GEF) 18
|
||||||
| Protein Structure | |||||||
| Useful resources | Stemformatics EHFPI ImmGen | ||||||
| Entrez Gene | |||||||
| Summary |
This gene does not have any Entrez summary - the following is the summary from its human ortholog ENSG00000104880:
Rho GTPases are GTP binding proteins that regulate a wide spectrum of cellular functions. These cellular processes include cytoskeletal rearrangements, gene transcription, cell growth and motility. Activation of Rho GTPases is under the direct control of guanine nucleotide exchange factors (GEFs). The protein encoded by this gene is a guanine nucleotide exchange factor and belongs to the Rho GTPase GFE family. Family members share a common feature, a Dbl (DH) homology domain followed by a pleckstrin (PH) homology domain. Alternatively spliced transcript variants encoding different isoforms have been identified. [provided by RefSeq, Oct 2008] |
||||||
| Gene Information | |||||||
| Type | Protein coding | ||||||
| Genomic Location | Chromosome 7:17513881-17565624 | ||||||
| Strand | Forward strand | ||||||
| Band | |||||||
| Transcripts |
|
||||||
| Interactions | |||||||
| Number of Interactions |
This gene and/or its encoded proteins are associated with 0 experimentally validated interaction(s) in this database.
They are also associated with 5 interaction(s) predicted by orthology.
|
||||||
| Gene Ontology | |||||||
Molecular Function |
|
||||||
| Biological Process |
|
||||||
| Cellular Component |
|
||||||
| Orthologs | |||||||
|
Species
Homo sapiens
Mus musculus
|
Gene ID
Gene Order
|
||||||
| Pathway Predictions based on Human Orthology Data | |||||||
| NETPATH | |||||||
| REACTOME |
NRAGE signals death through JNK pathway
G alpha (12/13) signalling events pathway
Rho GTPase cycle pathway
TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) pathway
SMAD4 MH2 Domain Mutants in Cancer pathway
Signalling by NGF pathway
Loss of Function of SMAD4 in Cancer pathway
Loss of Function of SMAD2/3 in Cancer pathway
Signaling by TGF-beta Receptor Complex in Cancer pathway
Signaling by Rho GTPases pathway
Signaling by GPCR pathway
p75 NTR receptor-mediated signalling pathway
TGFBR2 MSI Frameshift Mutants in Cancer pathway
Signal Transduction pathway
GPCR downstream signaling pathway
SMAD2/3 MH2 Domain Mutants in Cancer pathway
Loss of Function of TGFBR2 in Cancer pathway
SMAD2/3 Phosphorylation Motif Mutants in Cancer pathway
TGFBR1 KD Mutants in Cancer pathway
Cell death signalling via NRAGE, NRIF and NADE pathway
Loss of Function of TGFBR1 in Cancer pathway
TGFBR1 LBD Mutants in Cancer pathway
TGFBR2 Kinase Domain Mutants in Cancer pathway
Disease pathway
Signaling by TGF-beta Receptor Complex pathway
SMAD2/3 Phosphorylation Motif Mutants in Cancer pathway
GPCR downstream signaling pathway
Disease pathway
Signaling by GPCR pathway
Loss of Function of SMAD4 in Cancer pathway
NRAGE signals death through JNK pathway
Signaling by TGF-beta Receptor Complex in Cancer pathway
TGFBR1 LBD Mutants in Cancer pathway
Loss of Function of TGFBR2 in Cancer pathway
Signaling by Rho GTPases pathway
Cell death signalling via NRAGE, NRIF and NADE pathway
G alpha (12/13) signalling events pathway
TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) pathway
SMAD2/3 MH2 Domain Mutants in Cancer pathway
TGFBR2 Kinase Domain Mutants in Cancer pathway
Rho GTPase cycle pathway
Signalling by NGF pathway
Loss of Function of TGFBR1 in Cancer pathway
TGFBR2 MSI Frameshift Mutants in Cancer pathway
SMAD4 MH2 Domain Mutants in Cancer pathway
Signal Transduction pathway
TGFBR1 KD Mutants in Cancer pathway
p75 NTR receptor-mediated signalling pathway
Signaling by TGF-beta Receptor Complex pathway
Loss of Function of SMAD2/3 in Cancer pathway
|
||||||
| KEGG | |||||||
| INOH | |||||||
| PID NCI |
Regulation of RhoA activity
|
||||||
| Cross-References | |||||||
| SwissProt | |||||||
| TrEMBL | G3N3B9 | ||||||
| UniProt Splice Variant | |||||||
| Entrez Gene | |||||||
| UniGene | Bt.109942 Bt.111246 | ||||||
| RefSeq | |||||||
| HUGO | |||||||
| OMIM | |||||||
| CCDS | |||||||
| HPRD | |||||||
| IMGT | |||||||
| EMBL | DAAA02019498 | ||||||
| GenPept | |||||||
| RNA Seq Atlas | |||||||
| Transcript Frequencies | |||||||
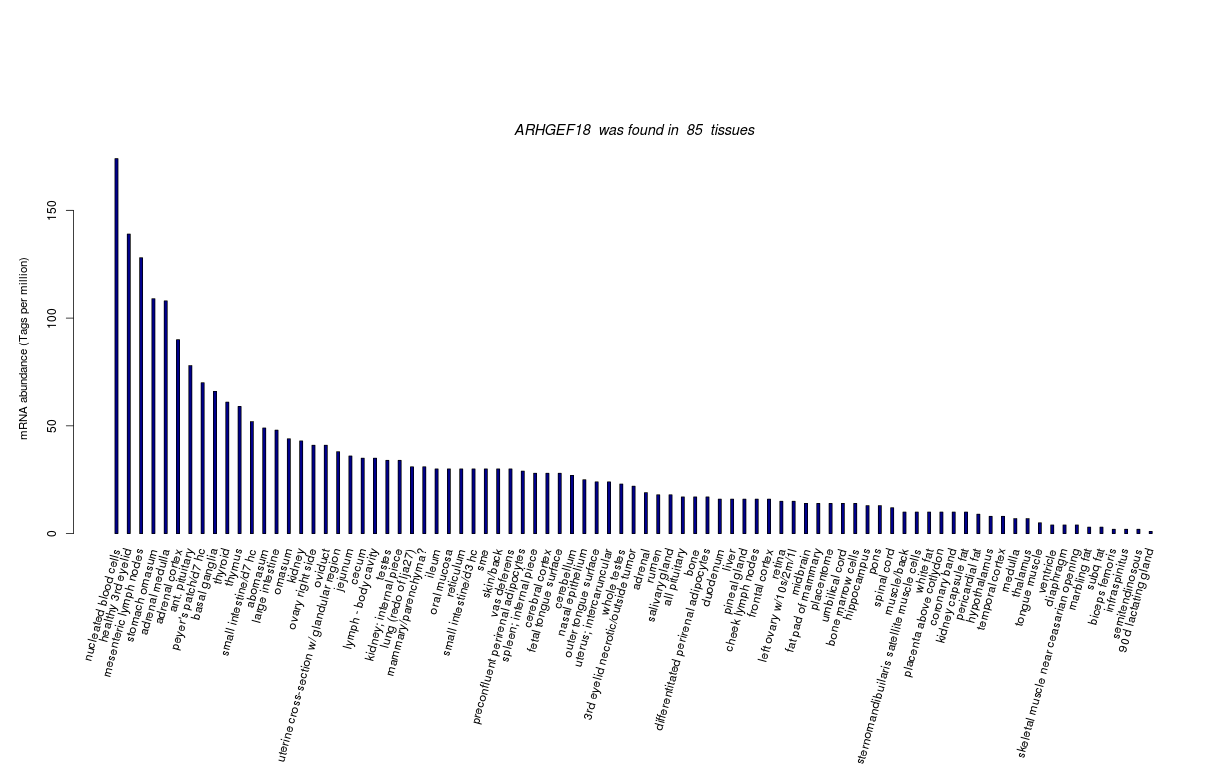
| Tag Count based mRNA-Abundances across 87 different Tissues (TPM).
Based on Data from Bovine Gene Atlas |
(Move your mouse over the image to view a more detailed version) |
||||||