| Bos taurus Gene: VAV1 | |||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Summary | |||||||||||||||||||||||||||||||||
| InnateDB Gene | IDBG-643888.3 | ||||||||||||||||||||||||||||||||
| Last Modified | 2014-10-13 [Report errors or provide feedback] | ||||||||||||||||||||||||||||||||
| Gene Symbol | VAV1 | ||||||||||||||||||||||||||||||||
| Gene Name | Proto-oncogene vav | ||||||||||||||||||||||||||||||||
| Synonyms | |||||||||||||||||||||||||||||||||
| Species | Bos taurus | ||||||||||||||||||||||||||||||||
| Ensembl Gene | ENSBTAG00000039160 | ||||||||||||||||||||||||||||||||
| Encoded Proteins |
vav 1 guanine nucleotide exchange factor
vav 1 guanine nucleotide exchange factor
|
||||||||||||||||||||||||||||||||
| Protein Structure | |||||||||||||||||||||||||||||||||
| Useful resources | Stemformatics EHFPI ImmGen | ||||||||||||||||||||||||||||||||
| Entrez Gene | |||||||||||||||||||||||||||||||||
| Summary |
This gene does not have any Entrez summary - the following is the summary from its human ortholog ENSG00000141968:
This gene is a member of the VAV gene family. The VAV proteins are guanine nucleotide exchange factors (GEFs) for Rho family GTPases that activate pathways leading to actin cytoskeletal rearrangements and transcriptional alterations. The encoded protein is important in hematopoiesis, playing a role in T-cell and B-cell development and activation. The encoded protein has been identified as the specific binding partner of Nef proteins from HIV-1. Coexpression and binding of these partners initiates profound morphological changes, cytoskeletal rearrangements and the JNK/SAPK signaling cascade, leading to increased levels of viral transcription and replication. Alternatively spliced transcript variants encoding multiple isoforms have been observed for this gene. [provided by RefSeq, Apr 2012] |
||||||||||||||||||||||||||||||||
| Gene Information | |||||||||||||||||||||||||||||||||
| Type | Protein coding | ||||||||||||||||||||||||||||||||
| Genomic Location | Chromosome 7:18866561-18937070 | ||||||||||||||||||||||||||||||||
| Strand | Reverse strand | ||||||||||||||||||||||||||||||||
| Band | |||||||||||||||||||||||||||||||||
| Transcripts |
|
||||||||||||||||||||||||||||||||
| Interactions | |||||||||||||||||||||||||||||||||
| Number of Interactions |
This gene and/or its encoded proteins are associated with 0 experimentally validated interaction(s) in this database.
They are also associated with 87 interaction(s) predicted by orthology.
|
||||||||||||||||||||||||||||||||
| Gene Ontology | |||||||||||||||||||||||||||||||||
Molecular Function |
|
||||||||||||||||||||||||||||||||
| Biological Process |
|
||||||||||||||||||||||||||||||||
| Cellular Component |
|
||||||||||||||||||||||||||||||||
| Orthologs | |||||||||||||||||||||||||||||||||
|
Species
Homo sapiens
Mus musculus
|
Gene ID
Gene Order
|
||||||||||||||||||||||||||||||||
| Pathway Predictions based on Human Orthology Data | |||||||||||||||||||||||||||||||||
| NETPATH |
EGFR1 pathway
KitReceptor pathway
TCR pathway
BCR pathway
IL2 pathway
IL3 pathway
IL5 pathway
IL6 pathway
Prolactin pathway
|
||||||||||||||||||||||||||||||||
| REACTOME |
PIP3 activates AKT signaling pathway
DAP12 signaling pathway
DAP12 interactions pathway
Regulation of actin dynamics for phagocytic cup formation pathway
Regulation of signaling by CBL pathway
Interleukin-3, 5 and GM-CSF signaling pathway
CD28 dependent Vav1 pathway pathway
CD28 co-stimulation pathway
Costimulation by the CD28 family pathway
Antigen activates B Cell Receptor (BCR) leading to generation of second messengers pathway
GPVI-mediated activation cascade pathway
PI3K/AKT activation pathway
NRAGE signals death through JNK pathway
GAB1 signalosome pathway
Signaling by EGFR pathway
G alpha (12/13) signalling events pathway
PI3K events in ERBB4 signaling pathway
Signaling by ERBB4 pathway
Rho GTPase cycle pathway
PI3K events in ERBB2 signaling pathway
Signaling by ERBB2 pathway
PI-3K cascade pathway
Signaling by FGFR pathway
Signaling by SCF-KIT pathway
Downstream signal transduction pathway
Signaling by PDGF pathway
Constitutive PI3K/AKT Signaling in Cancer pathway
Signaling by FGFR in disease pathway
Signalling by NGF pathway
Signaling by EGFR in Cancer pathway
Downstream signaling of activated FGFR pathway
Signaling by Rho GTPases pathway
Signaling by VEGF pathway
Role of LAT2/NTAL/LAB on calcium mobilization pathway
Cytokine Signaling in Immune system pathway
Signaling by GPCR pathway
Innate Immune System pathway
Platelet activation, signaling and aggregation pathway
VEGFR2 mediated vascular permeability pathway
Fc epsilon receptor (FCERI) signaling pathway
p75 NTR receptor-mediated signalling pathway
Signal Transduction pathway
GPCR downstream signaling pathway
Adaptive Immune System pathway
Immune System pathway
Signaling by Interleukins pathway
VEGFA-VEGFR2 Pathway pathway
NGF signalling via TRKA from the plasma membrane pathway
Fcgamma receptor (FCGR) dependent phagocytosis pathway
Cell death signalling via NRAGE, NRIF and NADE pathway
PI3K/AKT Signaling in Cancer pathway
FCERI mediated Ca+2 mobilization pathway
Signaling by the B Cell Receptor (BCR) pathway
Downstream signaling events of B Cell Receptor (BCR) pathway
Disease pathway
FCERI mediated MAPK activation pathway
Hemostasis pathway
Innate Immune System pathway
Regulation of actin dynamics for phagocytic cup formation pathway
Signaling by the B Cell Receptor (BCR) pathway
Signaling by ERBB2 pathway
Antigen activates B Cell Receptor (BCR) leading to generation of second messengers pathway
GPCR downstream signaling pathway
PI3K/AKT activation pathway
Disease pathway
Hemostasis pathway
Signaling by GPCR pathway
Signaling by VEGF pathway
Signaling by FGFR in disease pathway
Signaling by ERBB4 pathway
Signaling by SCF-KIT pathway
Cytokine Signaling in Immune system pathway
NRAGE signals death through JNK pathway
PIP3 activates AKT signaling pathway
Immune System pathway
Platelet activation, signaling and aggregation pathway
Downstream signal transduction pathway
Downstream signaling of activated FGFR pathway
Constitutive PI3K/AKT Signaling in Cancer pathway
PI3K events in ERBB4 signaling pathway
PI-3K cascade pathway
Signaling by Rho GTPases pathway
Cell death signalling via NRAGE, NRIF and NADE pathway
Signaling by EGFR pathway
GPVI-mediated activation cascade pathway
CD28 dependent Vav1 pathway pathway
G alpha (12/13) signalling events pathway
CD28 co-stimulation pathway
Costimulation by the CD28 family pathway
Role of LAT2/NTAL/LAB on calcium mobilization pathway
VEGFR2 mediated vascular permeability pathway
VEGFA-VEGFR2 Pathway pathway
Fc epsilon receptor (FCERI) signaling pathway
FCERI mediated Ca+2 mobilization pathway
Rho GTPase cycle pathway
Signaling by Interleukins pathway
PI3K/AKT Signaling in Cancer pathway
Fcgamma receptor (FCGR) dependent phagocytosis pathway
Signalling by NGF pathway
Adaptive Immune System pathway
Signaling by PDGF pathway
GAB1 signalosome pathway
NGF signalling via TRKA from the plasma membrane pathway
Interleukin-3, 5 and GM-CSF signaling pathway
PI3K events in ERBB2 signaling pathway
Signal Transduction pathway
p75 NTR receptor-mediated signalling pathway
Signaling by FGFR pathway
FCERI mediated MAPK activation pathway
Signaling by EGFR in Cancer pathway
Regulation of signaling by CBL pathway
Downstream signaling events of B Cell Receptor (BCR) pathway
DAP12 interactions pathway
DAP12 signaling pathway
|
||||||||||||||||||||||||||||||||
| KEGG |
Regulation of actin cytoskeleton pathway
Fc epsilon RI signaling pathway pathway
Leukocyte transendothelial migration pathway
B cell receptor signaling pathway pathway
Focal adhesion pathway
T cell receptor signaling pathway pathway
Natural killer cell mediated cytotoxicity pathway
Fc gamma R-mediated phagocytosis pathway
Chemokine signaling pathway pathway
T cell receptor signaling pathway pathway
Fc epsilon RI signaling pathway pathway
Natural killer cell mediated cytotoxicity pathway
Regulation of actin cytoskeleton pathway
Leukocyte transendothelial migration pathway
Focal adhesion pathway
B cell receptor signaling pathway pathway
Fc gamma R-mediated phagocytosis pathway
Chemokine signaling pathway pathway
|
||||||||||||||||||||||||||||||||
| INOH |
CD4 T cell receptor signaling pathway
B cell receptor signaling pathway
Integrin signaling pathway pathway
CD4 T cell receptor signaling pathway
B cell receptor signaling pathway
Integrin signaling pathway pathway
|
||||||||||||||||||||||||||||||||
| PID NCI |
TCR signaling in na´ve CD8+ T cells
Regulation of RAC1 activity
CXCR4-mediated signaling events
TCR signaling in na´ve CD4+ T cells
Regulation of RhoA activity
Signaling events mediated by Stem cell factor receptor (c-Kit)
Fc-epsilon receptor I signaling in mast cells
IL6-mediated signaling events
|
||||||||||||||||||||||||||||||||
| Cross-References | |||||||||||||||||||||||||||||||||
| SwissProt | |||||||||||||||||||||||||||||||||
| TrEMBL | F1MR20 | ||||||||||||||||||||||||||||||||
| UniProt Splice Variant | |||||||||||||||||||||||||||||||||
| Entrez Gene | |||||||||||||||||||||||||||||||||
| UniGene | |||||||||||||||||||||||||||||||||
| RefSeq | |||||||||||||||||||||||||||||||||
| HUGO | HGNC:12657 | ||||||||||||||||||||||||||||||||
| OMIM | |||||||||||||||||||||||||||||||||
| CCDS | |||||||||||||||||||||||||||||||||
| HPRD | |||||||||||||||||||||||||||||||||
| IMGT | |||||||||||||||||||||||||||||||||
| EMBL | DAAA02019536 DAAA02019537 DAAA02019538 DAAA02019539 DAAA02019540 DAAA02019541 DAAA02019542 DAAA02019543 DAAA02019544 | ||||||||||||||||||||||||||||||||
| GenPept | |||||||||||||||||||||||||||||||||
| RNA Seq Atlas | |||||||||||||||||||||||||||||||||
| Transcript Frequencies | |||||||||||||||||||||||||||||||||
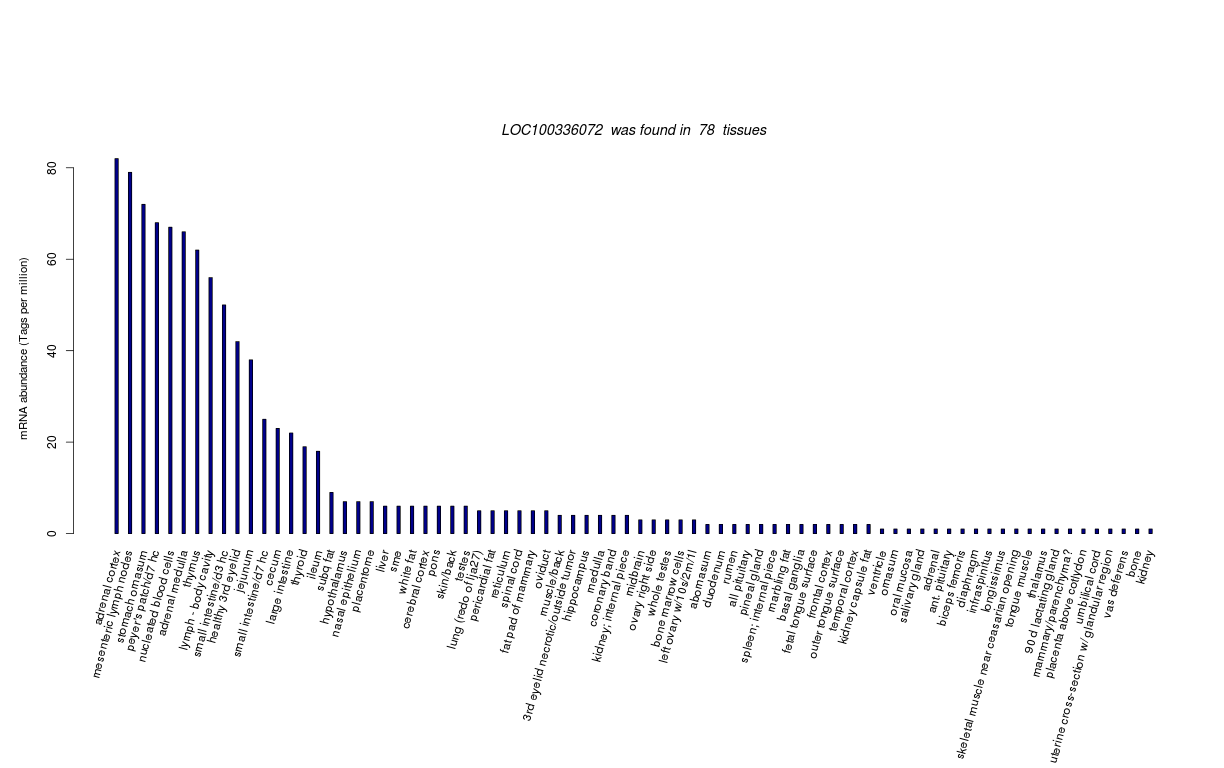
| Tag Count based mRNA-Abundances across 87 different Tissues (TPM).
Based on Data from Bovine Gene Atlas |
(Move your mouse over the image to view a more detailed version) |
||||||||||||||||||||||||||||||||