| Bos taurus Gene: PTP4A3 | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Summary | |||||||||||||
| InnateDB Gene | IDBG-643907.3 | ||||||||||||
| Last Modified | 2014-10-13 [Report errors or provide feedback] | ||||||||||||
| Gene Symbol | PTP4A3 | ||||||||||||
| Gene Name | Protein tyrosine phosphatase type IVA 3 | ||||||||||||
| Synonyms | |||||||||||||
| Species | Bos taurus | ||||||||||||
| Ensembl Gene | ENSBTAG00000046467 | ||||||||||||
| Encoded Proteins |
Protein tyrosine phosphatase type IVA 3
|
||||||||||||
| Protein Structure | |||||||||||||
| Useful resources | Stemformatics EHFPI ImmGen | ||||||||||||
| Entrez Gene | |||||||||||||
| Summary |
This gene does not have any Entrez summary - the following is the summary from its human ortholog ENSG00000184489:
The protein encoded by this gene belongs to a small class of prenylated protein tyrosine phosphatases (PTPs). PTPs are cell signaling molecules that play regulatory roles in a variety of cellular processes. This class of PTPs contain a PTP domain and a characteristic C-terminal prenylation motif. Studies of this class of PTPs in mice demonstrated that they were prenylated proteins in vivo, which suggested their association with cell plasma membrane. Overexpression of this gene in mammalian cells was reported to inhibit angiotensin-II induced cell calcium mobilization and promote cell growth. Two alternatively spliced variants exist. [provided by RefSeq, Jul 2008] This gene encodes a member of the protein-tyrosine phosphatase family. Protein tyrosine phosphatases are cell signaling molecules that play regulatory roles in a variety of cellular processes. Studies of this class of protein tyrosine phosphatase in mice demonstrates that they are prenylated in vivo, suggesting their association with cell plasma membrane. The encoded protein may enhance cell proliferation, and overexpression of this gene has been implicated in tumor metastasis. Alternative splicing results in multiple transcript variants. [provided by RefSeq, Jul 2013] |
||||||||||||
| Gene Information | |||||||||||||
| Type | Protein coding | ||||||||||||
| Genomic Location | Chromosome 14:3578346-3586015 | ||||||||||||
| Strand | Reverse strand | ||||||||||||
| Band | |||||||||||||
| Transcripts |
|
||||||||||||
| Interactions | |||||||||||||
| Number of Interactions |
This gene and/or its encoded proteins are associated with 0 experimentally validated interaction(s) in this database.
They are also associated with 94 interaction(s) predicted by orthology.
|
||||||||||||
| Gene Ontology | |||||||||||||
Molecular Function |
|
||||||||||||
| Biological Process |
|
||||||||||||
| Cellular Component |
|
||||||||||||
| Orthologs | |||||||||||||
|
Species
Homo sapiens
Mus musculus
|
Gene ID
Gene Order
|
||||||||||||
| Pathway Predictions based on Human Orthology Data | |||||||||||||
| NETPATH | |||||||||||||
| REACTOME | |||||||||||||
| KEGG | |||||||||||||
| INOH | |||||||||||||
| PID NCI |
Signaling events mediated by PRL
|
||||||||||||
| Cross-References | |||||||||||||
| SwissProt | |||||||||||||
| TrEMBL | |||||||||||||
| UniProt Splice Variant | |||||||||||||
| Entrez Gene | |||||||||||||
| UniGene | Bt.101302 | ||||||||||||
| RefSeq | NM_001083766 XM_005215123 XM_005215124 | ||||||||||||
| HUGO | |||||||||||||
| OMIM | |||||||||||||
| CCDS | |||||||||||||
| HPRD | |||||||||||||
| IMGT | |||||||||||||
| EMBL | |||||||||||||
| GenPept | |||||||||||||
| RNA Seq Atlas | |||||||||||||
| Transcript Frequencies | |||||||||||||
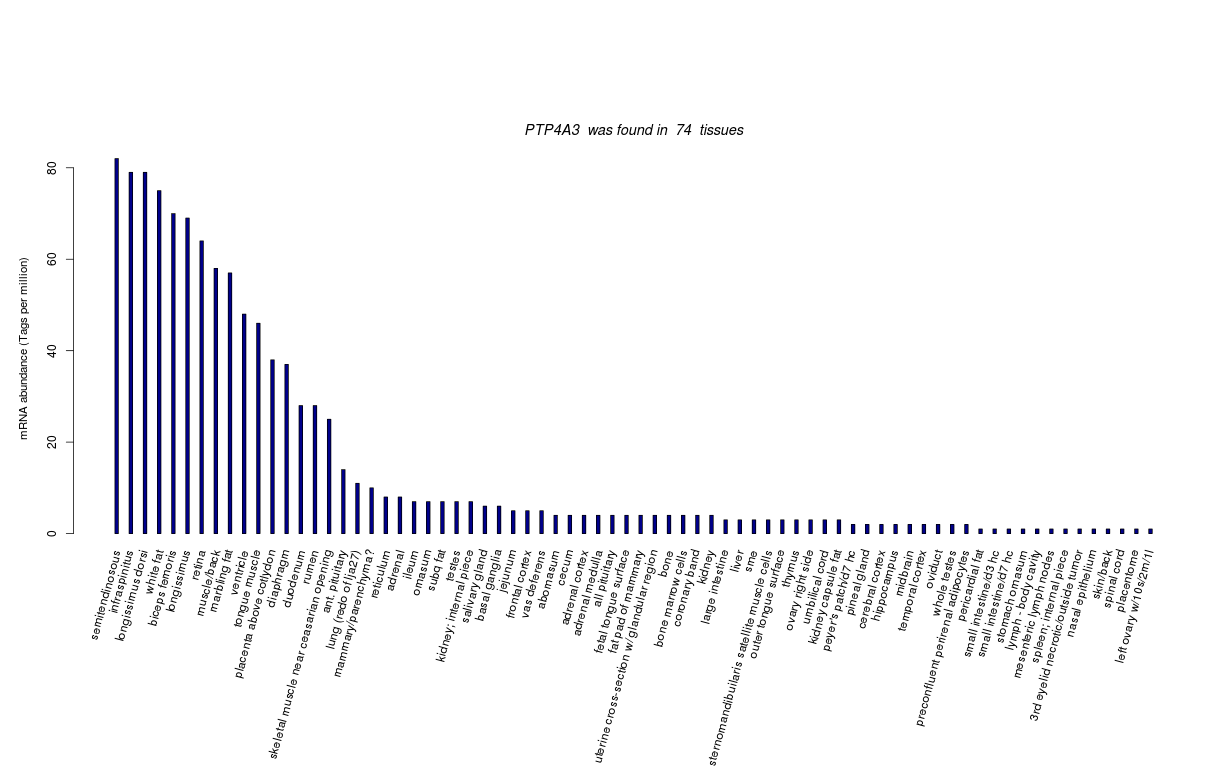
| Tag Count based mRNA-Abundances across 87 different Tissues (TPM).
Based on Data from Bovine Gene Atlas |
(Move your mouse over the image to view a more detailed version) |
||||||||||||