| Bos taurus Gene: PABPC1 | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Summary | |||||||||||||||||||
| InnateDB Gene | IDBG-645107.3 | ||||||||||||||||||
| Last Modified | 2014-10-13 [Report errors or provide feedback] | ||||||||||||||||||
| Gene Symbol | PABPC1 | ||||||||||||||||||
| Gene Name | Polyadenylate-binding protein 1 | ||||||||||||||||||
| Synonyms | |||||||||||||||||||
| Species | Bos taurus | ||||||||||||||||||
| Ensembl Gene | ENSBTAG00000046358 | ||||||||||||||||||
| Encoded Proteins |
Polyadenylate-binding protein 1
|
||||||||||||||||||
| Protein Structure |

|
||||||||||||||||||
| Useful resources | Stemformatics EHFPI ImmGen | ||||||||||||||||||
| Entrez Gene | |||||||||||||||||||
| Summary |
This gene does not have any Entrez summary - the following is the summary from its human ortholog ENSG00000070756:
This gene encodes a poly(A) binding protein. The protein shuttles between the nucleus and cytoplasm and binds to the 3' poly(A) tail of eukaryotic messenger RNAs via RNA-recognition motifs. The binding of this protein to poly(A) promotes ribosome recruitment and translation initiation; it is also required for poly(A) shortening which is the first step in mRNA decay. The gene is part of a small gene family including three protein-coding genes and several pseudogenes.[provided by RefSeq, Aug 2010] |
||||||||||||||||||
| Gene Information | |||||||||||||||||||
| Type | Protein coding | ||||||||||||||||||
| Genomic Location | Chromosome 14:65816006-65833756 | ||||||||||||||||||
| Strand | Forward strand | ||||||||||||||||||
| Band | |||||||||||||||||||
| Transcripts |
|
||||||||||||||||||
| Interactions | |||||||||||||||||||
| Number of Interactions |
This gene and/or its encoded proteins are associated with 0 experimentally validated interaction(s) in this database.
They are also associated with 141 interaction(s) predicted by orthology.
|
||||||||||||||||||
| Gene Ontology | |||||||||||||||||||
Molecular Function |
|
||||||||||||||||||
| Biological Process |
|
||||||||||||||||||
| Cellular Component |
|
||||||||||||||||||
| Orthologs | |||||||||||||||||||
|
Species
Homo sapiens
Mus musculus
|
Gene ID
Gene Order
|
||||||||||||||||||
| Pathways | |||||||||||||||||||
| NETPATH | |||||||||||||||||||
| REACTOME |
Deadenylation of mRNA pathway
Regulation of mRNA stability by proteins that bind AU-rich elements pathway
L13a-mediated translational silencing of Ceruloplasmin expression pathway
Nonsense Mediated Decay (NMD) enhanced by the Exon Junction Complex (EJC) pathway
Gene Expression pathway
Eukaryotic Translation Initiation pathway
Cap-dependent Translation Initiation pathway
Nonsense Mediated Decay (NMD) independent of the Exon Junction Complex (EJC) pathway
AUF1 (hnRNP D0) destabilizes mRNA pathway
Translation pathway
Metabolism of proteins pathway
Activation of the mRNA upon binding of the cap-binding complex and eIFs, and subsequent binding to 43S pathway
Translation initiation complex formation pathway
Nonsense-Mediated Decay (NMD) pathway
Deadenylation-dependent mRNA decay pathway
|
||||||||||||||||||
| KEGG | |||||||||||||||||||
| INOH | |||||||||||||||||||
| PID NCI | |||||||||||||||||||
| Pathway Predictions based on Human Orthology Data | |||||||||||||||||||
| NETPATH |
TSLP pathway
|
||||||||||||||||||
| REACTOME |
L13a-mediated translational silencing of Ceruloplasmin expression pathway
AUF1 (hnRNP D0) destabilizes mRNA pathway
Deadenylation of mRNA pathway
Nonsense Mediated Decay (NMD) independent of the Exon Junction Complex (EJC) pathway
Nonsense Mediated Decay (NMD) enhanced by the Exon Junction Complex (EJC) pathway
Translation initiation complex formation pathway
Activation of the mRNA upon binding of the cap-binding complex and eIFs, and subsequent binding to 43S pathway
Eukaryotic Translation Initiation pathway
Nonsense-Mediated Decay (NMD) pathway
Deadenylation-dependent mRNA decay pathway
Translation pathway
Metabolism of proteins pathway
Cap-dependent Translation Initiation pathway
Regulation of mRNA stability by proteins that bind AU-rich elements pathway
Gene Expression pathway
Activation of the mRNA upon binding of the cap-binding complex and eIFs, and subsequent binding to 43S pathway
Regulation of mRNA stability by proteins that bind AU-rich elements pathway
AUF1 (hnRNP D0) destabilizes mRNA pathway
Deadenylation of mRNA pathway
Translation pathway
Translation initiation complex formation pathway
Nonsense-Mediated Decay (NMD) pathway
Metabolism of proteins pathway
Nonsense Mediated Decay (NMD) enhanced by the Exon Junction Complex (EJC) pathway
Nonsense Mediated Decay (NMD) independent of the Exon Junction Complex (EJC) pathway
Cap-dependent Translation Initiation pathway
Eukaryotic Translation Initiation pathway
Gene Expression pathway
L13a-mediated translational silencing of Ceruloplasmin expression pathway
Deadenylation-dependent mRNA decay pathway
|
||||||||||||||||||
| KEGG |
RNA degradation pathway
RNA transport pathway
mRNA surveillance pathway pathway
RNA degradation pathway
RNA transport pathway
mRNA surveillance pathway pathway
|
||||||||||||||||||
| INOH | |||||||||||||||||||
| PID NCI | |||||||||||||||||||
| Cross-References | |||||||||||||||||||
| SwissProt | P61286 | ||||||||||||||||||
| TrEMBL | |||||||||||||||||||
| UniProt Splice Variant | |||||||||||||||||||
| Entrez Gene | 282296 | ||||||||||||||||||
| UniGene | Bt.111653 Bt.53579 Bt.89769 | ||||||||||||||||||
| RefSeq | NM_174568 | ||||||||||||||||||
| HUGO | |||||||||||||||||||
| OMIM | |||||||||||||||||||
| CCDS | |||||||||||||||||||
| HPRD | |||||||||||||||||||
| IMGT | |||||||||||||||||||
| EMBL | AJ401269 BC102365 | ||||||||||||||||||
| GenPept | AAI02366 CAB96752 | ||||||||||||||||||
| RNA Seq Atlas | 282296 | ||||||||||||||||||
| Transcript Frequencies | |||||||||||||||||||
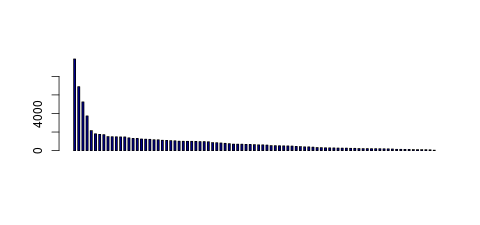
| Tag Count based mRNA-Abundances across 87 different Tissues (TPM).
Based on Data from Bovine Gene Atlas |
(Move your mouse over the image to view a more detailed version) |
||||||||||||||||||