| Bos taurus Gene: NRAS | |||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Summary | |||||||||||||||||||||||||||||||
| InnateDB Gene | IDBG-634702.3 | ||||||||||||||||||||||||||||||
| Last Modified | 2014-10-13 [Report errors or provide feedback] | ||||||||||||||||||||||||||||||
| Gene Symbol | NRAS | ||||||||||||||||||||||||||||||
| Gene Name | GTPase NRas | ||||||||||||||||||||||||||||||
| Synonyms | |||||||||||||||||||||||||||||||
| Species | Bos taurus | ||||||||||||||||||||||||||||||
| Ensembl Gene | ENSBTAG00000046797 | ||||||||||||||||||||||||||||||
| Encoded Proteins |
GTPase NRas
|
||||||||||||||||||||||||||||||
| Protein Structure | |||||||||||||||||||||||||||||||
| Useful resources | Stemformatics EHFPI ImmGen | ||||||||||||||||||||||||||||||
| InnateDB Annotation from Orthologs | |||||||||||||||||||||||||||||||
| Summary |
[Mus musculus] Nras is a small GTPase that, in response to microbial activation, mediates cholangiocyte pro-inflammatory cytokine production and induction of cholangiocyte proliferation. (Demonstrated in human)
[Homo sapiens] NRAS is a small GTPase that, in response to microbial activation, mediates cholangiocyte pro-inflammatory cytokine production and induction of cholangiocyte proliferation.
|
||||||||||||||||||||||||||||||
| Entrez Gene | |||||||||||||||||||||||||||||||
| Summary |
This gene does not have any Entrez summary - the following is the summary from its human ortholog ENSG00000213281:
This is an N-ras oncogene encoding a membrane protein that shuttles between the Golgi apparatus and the plasma membrane. This shuttling is regulated through palmitoylation and depalmitoylation by the ZDHHC9-GOLGA7 complex. The encoded protein, which has intrinsic GTPase activity, is activated by a guanine nucleotide-exchange factor and inactivated by a GTPase activating protein. Mutations in this gene have been associated with somatic rectal cancer, follicular thyroid cancer, autoimmune lymphoproliferative syndrome, Noonan syndrome, and juvenile myelomonocytic leukemia. [provided by RefSeq, Jun 2011] |
||||||||||||||||||||||||||||||
| Gene Information | |||||||||||||||||||||||||||||||
| Type | Protein coding | ||||||||||||||||||||||||||||||
| Genomic Location | Chromosome 3:28723874-28733770 | ||||||||||||||||||||||||||||||
| Strand | Forward strand | ||||||||||||||||||||||||||||||
| Band | |||||||||||||||||||||||||||||||
| Transcripts |
|
||||||||||||||||||||||||||||||
| Interactions | |||||||||||||||||||||||||||||||
| Number of Interactions |
This gene and/or its encoded proteins are associated with 0 experimentally validated interaction(s) in this database.
They are also associated with 26 interaction(s) predicted by orthology.
|
||||||||||||||||||||||||||||||
| Gene Ontology | |||||||||||||||||||||||||||||||
Molecular Function |
|
||||||||||||||||||||||||||||||
| Biological Process |
|
||||||||||||||||||||||||||||||
| Cellular Component |
|
||||||||||||||||||||||||||||||
| Orthologs | |||||||||||||||||||||||||||||||
|
Species
Mus musculus
Homo sapiens
|
Gene ID
Gene Order
|
||||||||||||||||||||||||||||||
| Pathway Predictions based on Human Orthology Data | |||||||||||||||||||||||||||||||
| NETPATH | |||||||||||||||||||||||||||||||
| REACTOME |
RAF activation pathway
RAF phosphorylates MEK pathway
MEK activation pathway
DAP12 signaling pathway
DAP12 interactions pathway
Interleukin-2 signaling pathway
Activation of RAS in B cells pathway
Tie2 Signaling pathway
Cell surface interactions at the vascular wall pathway
Frs2-mediated activation pathway
ARMS-mediated activation pathway
p38MAPK events pathway
Signalling to p38 via RIT and RIN pathway
Signalling to RAS pathway
SOS-mediated signalling pathway
SHC1 events in EGFR signaling pathway
GRB2 events in EGFR signaling pathway
Signaling by EGFR pathway
EGFR Transactivation by Gastrin pathway
Gastrin-CREB signalling pathway via PKC and MAPK pathway
SHC1 events in ERBB4 signaling pathway
Signaling by ERBB4 pathway
SHC-mediated signalling pathway
Signaling by Insulin receptor pathway
IRS-related events triggered by IGF1R pathway
SHC-related events triggered by IGF1R pathway
Signaling by Type 1 Insulin-like Growth Factor 1 Receptor (IGF1R) pathway
GRB2 events in ERBB2 signaling pathway
SHC1 events in ERBB2 signaling pathway
Signaling by ERBB2 pathway
SHC-mediated cascade pathway
FRS2-mediated cascade pathway
Signaling by FGFR pathway
Signaling by SCF-KIT pathway
Downstream signal transduction pathway
Signaling by PDGF pathway
Signaling by constitutively active EGFR pathway
Signaling by FGFR mutants pathway
NCAM signaling for neurite out-growth pathway
Signaling by FGFR in disease pathway
SHC-related events pathway
Developmental Biology pathway
Signalling by NGF pathway
Signaling by EGFR in Cancer pathway
Downstream signaling of activated FGFR pathway
Signaling by VEGF pathway
Cytokine Signaling in Immune system pathway
Signaling by GPCR pathway
Innate Immune System pathway
Axon guidance pathway
RAF/MAP kinase cascade pathway
Fc epsilon receptor (FCERI) signaling pathway
Signal Transduction pathway
IRS-mediated signalling pathway
Adaptive Immune System pathway
Immune System pathway
VEGFR2 mediated cell proliferation pathway
Insulin receptor signalling cascade pathway
Signaling by Interleukins pathway
VEGFA-VEGFR2 Pathway pathway
Signalling to ERKs pathway
NGF signalling via TRKA from the plasma membrane pathway
IRS-related events pathway
IGF1R signaling cascade pathway
Prolonged ERK activation events pathway
Signaling by the B Cell Receptor (BCR) pathway
Downstream signaling events of B Cell Receptor (BCR) pathway
Disease pathway
Signaling by Leptin pathway
FCERI mediated MAPK activation pathway
Hemostasis pathway
|
||||||||||||||||||||||||||||||
| KEGG |
Tight junction pathway
Endometrial cancer pathway
Axon guidance pathway
Bladder cancer pathway
Renal cell carcinoma pathway
ErbB signaling pathway pathway
VEGF signaling pathway pathway
Insulin signaling pathway pathway
Acute myeloid leukemia pathway
T cell receptor signaling pathway pathway
Melanogenesis pathway
Gap junction pathway
Thyroid cancer pathway
Fc epsilon RI signaling pathway pathway
Melanoma pathway
Natural killer cell mediated cytotoxicity pathway
GnRH signaling pathway pathway
Regulation of actin cytoskeleton pathway
MAPK signaling pathway pathway
Long-term potentiation pathway
Glioma pathway
Prostate cancer pathway
Long-term depression pathway
Chronic myeloid leukemia pathway
Non-small cell lung cancer pathway
B cell receptor signaling pathway pathway
Chemokine signaling pathway pathway
Neurotrophin signaling pathway pathway
Pathways in cancer pathway
Hepatitis C pathway
Regulation of actin cytoskeleton pathway
GnRH signaling pathway pathway
Acute myeloid leukemia pathway
VEGF signaling pathway pathway
Fc epsilon RI signaling pathway pathway
Gap junction pathway
Non-small cell lung cancer pathway
ErbB signaling pathway pathway
Renal cell carcinoma pathway
MAPK signaling pathway pathway
Melanoma pathway
Axon guidance pathway
Thyroid cancer pathway
Endometrial cancer pathway
Long-term potentiation pathway
Bladder cancer pathway
Tight junction pathway
Glioma pathway
Insulin signaling pathway pathway
B cell receptor signaling pathway pathway
Prostate cancer pathway
Long-term depression pathway
T cell receptor signaling pathway pathway
Natural killer cell mediated cytotoxicity pathway
Chronic myeloid leukemia pathway
Melanogenesis pathway
Neurotrophin signaling pathway pathway
Chemokine signaling pathway pathway
Pathways in cancer pathway
Hepatitis C pathway
|
||||||||||||||||||||||||||||||
| INOH |
CD4 T cell receptor signaling pathway
JAK STAT pathway and regulation pathway
B cell receptor signaling pathway
Integrin signaling pathway pathway
FGF signaling pathway pathway
Insulin receptor signaling pathway
HGF signaling pathway pathway
EGF signaling pathway pathway
NGF signaling pathway pathway
IGF1 signaling pathway pathway
PDGF signaling pathway pathway
|
||||||||||||||||||||||||||||||
| PID NCI |
C-MYB transcription factor network
Regulation of Ras family activation
|
||||||||||||||||||||||||||||||
| Cross-References | |||||||||||||||||||||||||||||||
| SwissProt | |||||||||||||||||||||||||||||||
| TrEMBL | |||||||||||||||||||||||||||||||
| UniProt Splice Variant | |||||||||||||||||||||||||||||||
| Entrez Gene | |||||||||||||||||||||||||||||||
| UniGene | Bt.53950 | ||||||||||||||||||||||||||||||
| RefSeq | NM_001097989 | ||||||||||||||||||||||||||||||
| HUGO | |||||||||||||||||||||||||||||||
| OMIM | |||||||||||||||||||||||||||||||
| CCDS | |||||||||||||||||||||||||||||||
| HPRD | |||||||||||||||||||||||||||||||
| IMGT | |||||||||||||||||||||||||||||||
| EMBL | |||||||||||||||||||||||||||||||
| GenPept | |||||||||||||||||||||||||||||||
| RNA Seq Atlas | |||||||||||||||||||||||||||||||
| Transcript Frequencies | |||||||||||||||||||||||||||||||
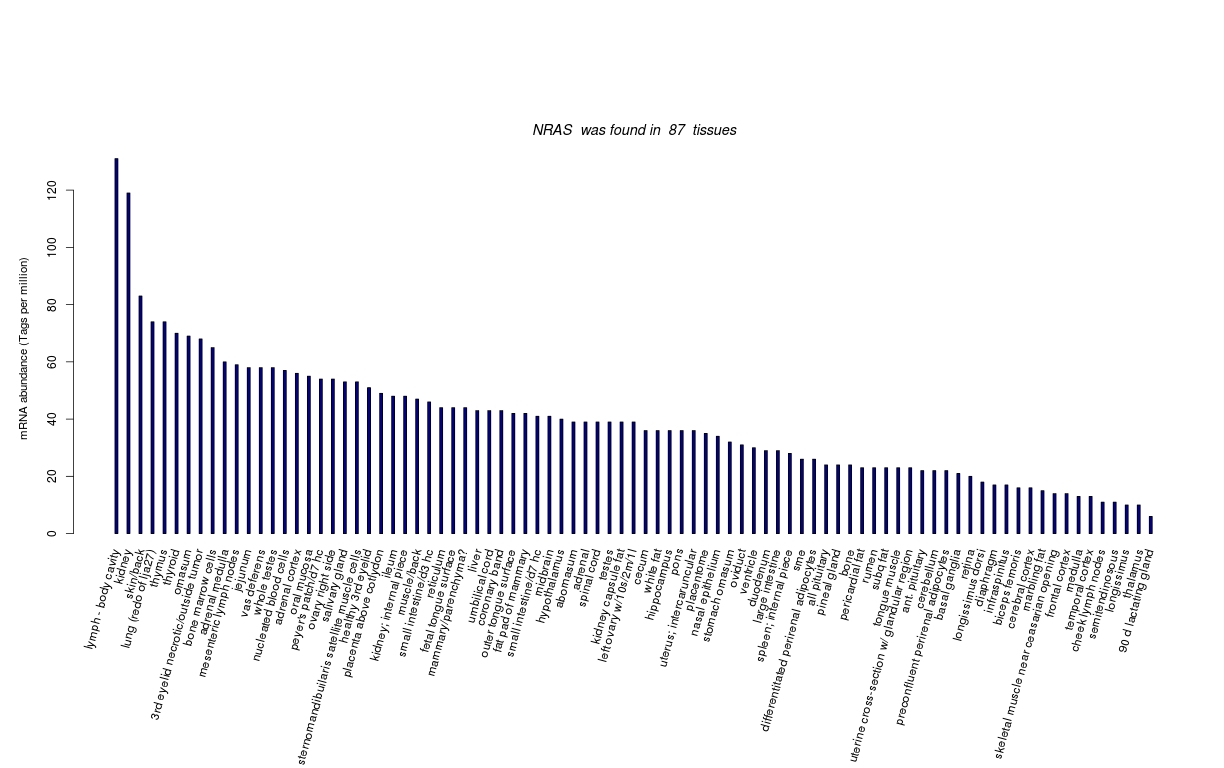
| Tag Count based mRNA-Abundances across 87 different Tissues (TPM).
Based on Data from Bovine Gene Atlas |
(Move your mouse over the image to view a more detailed version) |
||||||||||||||||||||||||||||||