| Bos taurus Gene: PDGFC | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Summary | |||||||||||||||||||
| InnateDB Gene | IDBG-630627.3 | ||||||||||||||||||
| Last Modified | 2014-10-13 [Report errors or provide feedback] | ||||||||||||||||||
| Gene Symbol | PDGFC | ||||||||||||||||||
| Gene Name | Uncharacterized protein | ||||||||||||||||||
| Synonyms | |||||||||||||||||||
| Species | Bos taurus | ||||||||||||||||||
| Ensembl Gene | ENSBTAG00000043959 | ||||||||||||||||||
| Encoded Proteins |
Uncharacterized protein
|
||||||||||||||||||
| Protein Structure | |||||||||||||||||||
| Useful resources | Stemformatics EHFPI ImmGen | ||||||||||||||||||
| Entrez Gene | |||||||||||||||||||
| Summary |
This gene does not have any Entrez summary - the following is the summary from its human ortholog ENSG00000145431:
The protein encoded by this gene is a member of the platelet-derived growth factor family. The four members of this family are mitogenic factors for cells of mesenchymal origin and are characterized by a core motif of eight cysteines. This gene product appears to form only homodimers. It differs from the platelet-derived growth factor alpha and beta polypeptides in having an unusual N-terminal domain, the CUB domain. Alternatively spliced transcript variants have been found for this gene. [provided by RefSeq, Sep 2010] |
||||||||||||||||||
| Gene Information | |||||||||||||||||||
| Type | Protein coding | ||||||||||||||||||
| Genomic Location | Chromosome 17:43348702-43457620 | ||||||||||||||||||
| Strand | Forward strand | ||||||||||||||||||
| Band | |||||||||||||||||||
| Transcripts |
|
||||||||||||||||||
| Interactions | |||||||||||||||||||
| Number of Interactions |
This gene and/or its encoded proteins are associated with 0 experimentally validated interaction(s) in this database.
They are also associated with 4 interaction(s) predicted by orthology.
|
||||||||||||||||||
| Gene Ontology | |||||||||||||||||||
Molecular Function |
|
||||||||||||||||||
| Biological Process |
|
||||||||||||||||||
| Cellular Component |
|
||||||||||||||||||
| Orthologs | |||||||||||||||||||
|
Species
Homo sapiens
Mus musculus
|
Gene ID
Gene Order
|
||||||||||||||||||
| Pathway Predictions based on Human Orthology Data | |||||||||||||||||||
| NETPATH | |||||||||||||||||||
| REACTOME |
Signaling by PDGF pathway
Signal Transduction pathway
|
||||||||||||||||||
| KEGG |
Regulation of actin cytoskeleton pathway
Gap junction pathway
Melanoma pathway
Cytokine-cytokine receptor interaction pathway
Prostate cancer pathway
Focal adhesion pathway
Cytokine-cytokine receptor interaction pathway
Gap junction pathway
Melanoma pathway
Regulation of actin cytoskeleton pathway
Prostate cancer pathway
Focal adhesion pathway
|
||||||||||||||||||
| INOH | |||||||||||||||||||
| PID NCI |
PDGF receptor signaling network
|
||||||||||||||||||
| Cross-References | |||||||||||||||||||
| SwissProt | |||||||||||||||||||
| TrEMBL | |||||||||||||||||||
| UniProt Splice Variant | |||||||||||||||||||
| Entrez Gene | |||||||||||||||||||
| UniGene | |||||||||||||||||||
| RefSeq | XM_002694441 XM_005194984 XM_005217687 XM_864899 | ||||||||||||||||||
| HUGO | |||||||||||||||||||
| OMIM | |||||||||||||||||||
| CCDS | |||||||||||||||||||
| HPRD | |||||||||||||||||||
| IMGT | |||||||||||||||||||
| EMBL | |||||||||||||||||||
| GenPept | |||||||||||||||||||
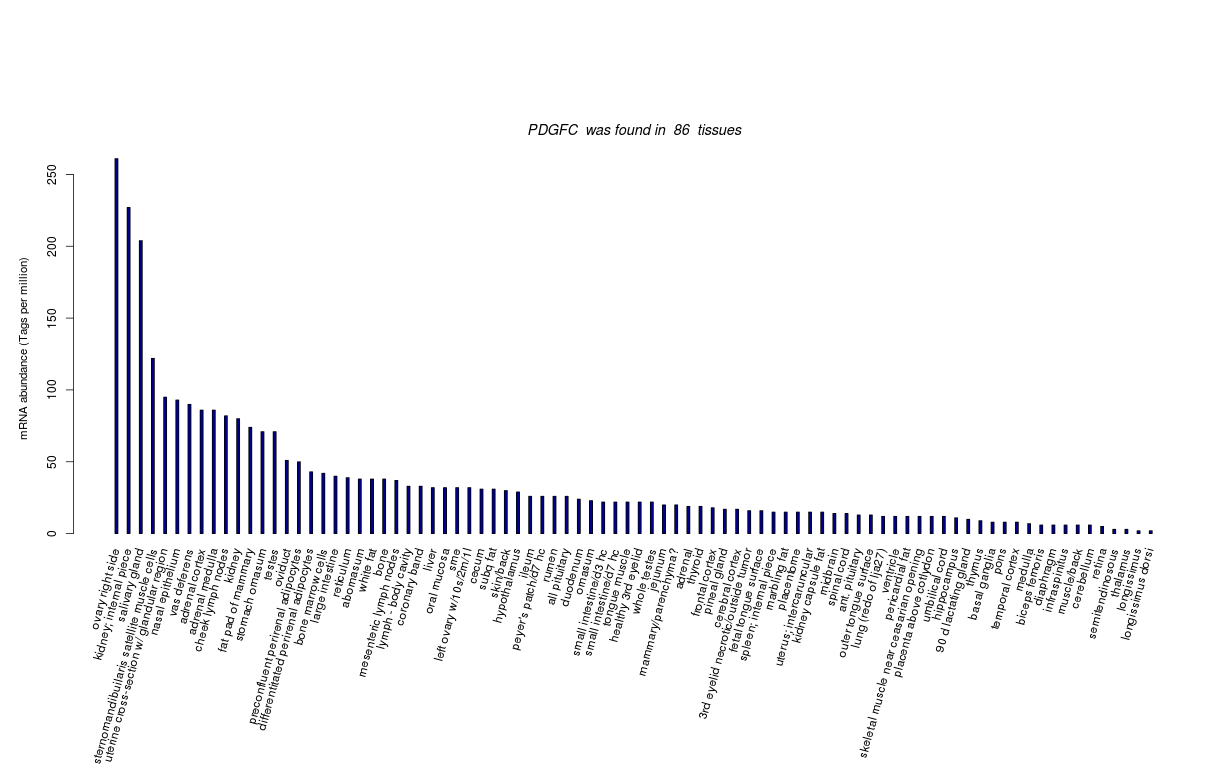
| RNA Seq Atlas | |||||||||||||||||||
| Transcript Frequencies | |||||||||||||||||||
| Tag Count based mRNA-Abundances across 87 different Tissues (TPM).
Based on Data from Bovine Gene Atlas |
(Move your mouse over the image to view a more detailed version) |
||||||||||||||||||