| Bos taurus Gene: BT.90254 | |||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Summary | |||||||||||||||||||||
| InnateDB Gene | IDBG-635691.3 | ||||||||||||||||||||
| Last Modified | 2014-10-13 [Report errors or provide feedback] | ||||||||||||||||||||
| Gene Symbol | BT.90254 | ||||||||||||||||||||
| Gene Name | histone deacetylase 8 | ||||||||||||||||||||
| Synonyms | |||||||||||||||||||||
| Species | Bos taurus | ||||||||||||||||||||
| Ensembl Gene | ENSBTAG00000046092 | ||||||||||||||||||||
| Encoded Proteins |
histone deacetylase 8
|
||||||||||||||||||||
| Protein Structure | |||||||||||||||||||||
| Useful resources | Stemformatics EHFPI ImmGen | ||||||||||||||||||||
| Entrez Gene | |||||||||||||||||||||
| Summary |
This gene does not have any Entrez summary - the following is the summary from its human ortholog ENSG00000147099:
Histones play a critical role in transcriptional regulation, cell cycle progression, and developmental events. Histone acetylation/deacetylation alters chromosome structure and affects transcription factor access to DNA. The protein encoded by this gene belongs to class I of the histone deacetylase family. It catalyzes the deacetylation of lysine residues in the histone N-terminal tails and represses transcription in large multiprotein complexes with transcriptional co-repressors. Multiple transcript variants encoding different isoforms have been found for this gene. [provided by RefSeq, Oct 2009] |
||||||||||||||||||||
| Gene Information | |||||||||||||||||||||
| Type | Protein coding | ||||||||||||||||||||
| Genomic Location | Chromosome X:83228463-83475076 | ||||||||||||||||||||
| Strand | Forward strand | ||||||||||||||||||||
| Band | |||||||||||||||||||||
| Transcripts |
|
||||||||||||||||||||
| Interactions | |||||||||||||||||||||
| Number of Interactions |
This gene and/or its encoded proteins are associated with 0 experimentally validated interaction(s) in this database.
They are also associated with 66 interaction(s) predicted by orthology.
|
||||||||||||||||||||
| Gene Ontology | |||||||||||||||||||||
Molecular Function |
|
||||||||||||||||||||
| Biological Process |
|
||||||||||||||||||||
| Cellular Component |
|
||||||||||||||||||||
| Orthologs | |||||||||||||||||||||
|
Species
Homo sapiens
Mus musculus
|
Gene ID
Gene Order
|
||||||||||||||||||||
| Pathway Predictions based on Human Orthology Data | |||||||||||||||||||||
| NETPATH | |||||||||||||||||||||
| REACTOME |
NOTCH1 Intracellular Domain Regulates Transcription pathway
Constitutive Signaling by NOTCH1 PEST Domain Mutants pathway
Constitutive Signaling by NOTCH1 HD+PEST Domain Mutants pathway
Signaling by NOTCH1 t(7;9)(NOTCH1:M1580_K2555) Translocation Mutant pathway
Signaling by NOTCH1 in Cancer pathway
FBXW7 Mutants and NOTCH1 in Cancer pathway
Signal Transduction pathway
Signaling by NOTCH1 PEST Domain Mutants in Cancer pathway
HDACs deacetylate histones pathway
Signaling by NOTCH1 HD+PEST Domain Mutants in Cancer pathway
Chromatin organization pathway
Signaling by NOTCH pathway
Chromatin modifying enzymes pathway
Signaling by NOTCH1 pathway
Signaling by NOTCH1 HD Domain Mutants in Cancer pathway
Disease pathway
Cell Cycle, Mitotic pathway
Chromatin organization pathway
Separation of Sister Chromatids pathway
Resolution of Sister Chromatid Cohesion pathway
Cell Cycle pathway
M Phase pathway
Chromatin modifying enzymes pathway
Mitotic Metaphase and Anaphase pathway
Mitotic Prometaphase pathway
HDACs deacetylate histones pathway
Mitotic Anaphase pathway
|
||||||||||||||||||||
| KEGG | |||||||||||||||||||||
| INOH |
TGF-beta signaling pathway
TGF-beta signaling pathway
|
||||||||||||||||||||
| PID NCI |
Signaling events mediated by HDAC Class I
|
||||||||||||||||||||
| Cross-References | |||||||||||||||||||||
| SwissProt | |||||||||||||||||||||
| TrEMBL | G3MYR9 | ||||||||||||||||||||
| UniProt Splice Variant | |||||||||||||||||||||
| Entrez Gene | 540666 | ||||||||||||||||||||
| UniGene | Bt.90254 | ||||||||||||||||||||
| RefSeq | NM_001076231 | ||||||||||||||||||||
| HUGO | |||||||||||||||||||||
| OMIM | |||||||||||||||||||||
| CCDS | |||||||||||||||||||||
| HPRD | |||||||||||||||||||||
| IMGT | |||||||||||||||||||||
| EMBL | DAAA02072645 DAAA02072646 DAAA02072647 DAAA02072648 DAAA02072649 DAAA02072650 DAAA02072651 DAAA02072652 DAAA02072653 DAAA02072654 DAAA02072655 DAAA02072656 | ||||||||||||||||||||
| GenPept | |||||||||||||||||||||
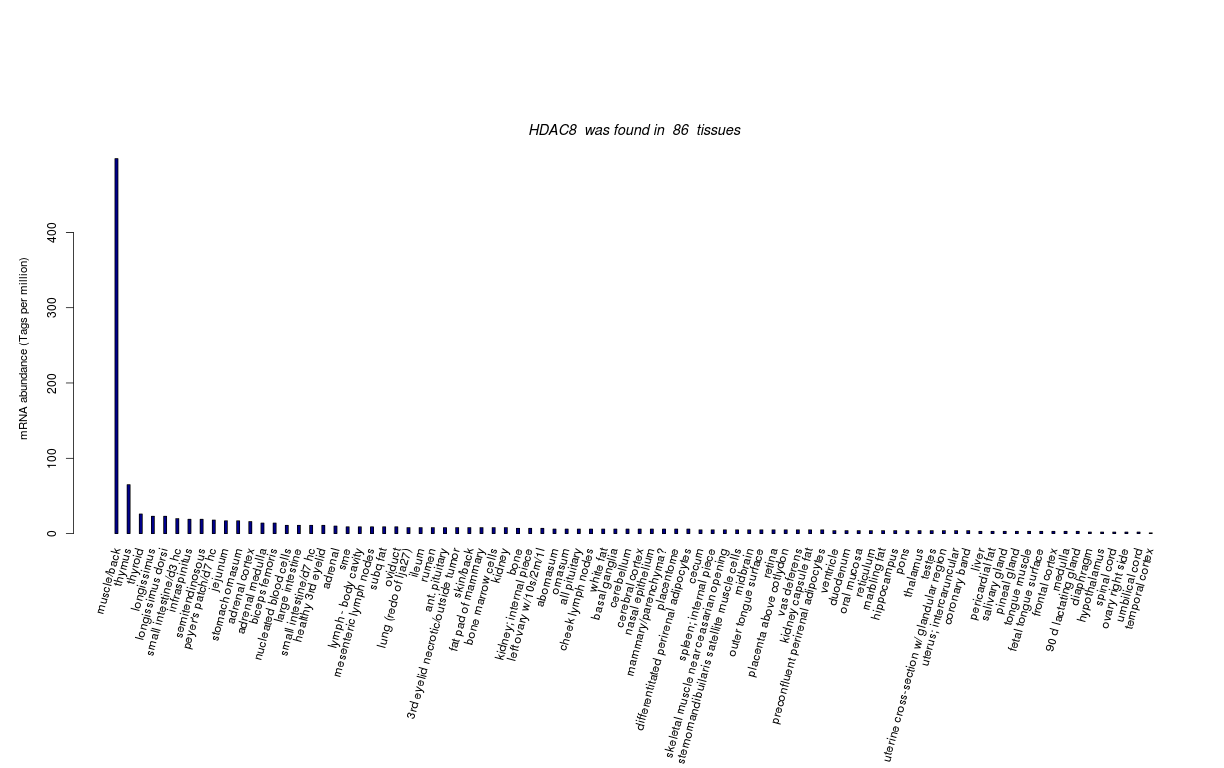
| RNA Seq Atlas | 540666 | ||||||||||||||||||||
| Transcript Frequencies | |||||||||||||||||||||
| Tag Count based mRNA-Abundances across 87 different Tissues (TPM).
Based on Data from Bovine Gene Atlas |
(Move your mouse over the image to view a more detailed version) |
||||||||||||||||||||