| Bos taurus Gene: LFNG | |||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Summary | |||||||||||||||||||||||
| InnateDB Gene | IDBG-641284.3 | ||||||||||||||||||||||
| Last Modified | 2014-10-13 [Report errors or provide feedback] | ||||||||||||||||||||||
| Gene Symbol | LFNG | ||||||||||||||||||||||
| Gene Name | Beta-1,3-N-acetylglucosaminyltransferase lunatic fringe | ||||||||||||||||||||||
| Synonyms | |||||||||||||||||||||||
| Species | Bos taurus | ||||||||||||||||||||||
| Ensembl Gene | ENSBTAG00000040361 | ||||||||||||||||||||||
| Encoded Proteins |
Beta-1,3-N-acetylglucosaminyltransferase lunatic fringe
|
||||||||||||||||||||||
| Protein Structure | |||||||||||||||||||||||
| Useful resources | Stemformatics EHFPI ImmGen | ||||||||||||||||||||||
| Entrez Gene | |||||||||||||||||||||||
| Summary |
This gene does not have any Entrez summary - the following is the summary from its human ortholog ENSG00000106003:
This gene is a member of the fringe gene family which also includes radical and manic fringe genes. They all encode evolutionarily conserved glycosyltransferases that act in the Notch signaling pathway to define boundaries during embryonic development. While their genomic structure is distinct from other glycosyltransferases, fringe proteins have a fucose-specific beta-1,3-N-acetylglucosaminyltransferase activity that leads to elongation of O-linked fucose residues on Notch, which alters Notch signaling. This gene product is predicted to be a single-pass type II Golgi membrane protein but it may also be secreted and proteolytically processed like the related proteins in mouse and Drosophila (PMID: 9187150). Mutations in this gene have been associated with autosomal recessive spondylocostal dysostosis 3. Multiple transcript variants encoding different isoforms have been found for this gene. [provided by RefSeq, Oct 2009] |
||||||||||||||||||||||
| Gene Information | |||||||||||||||||||||||
| Type | Protein coding | ||||||||||||||||||||||
| Genomic Location | Chromosome 25:41304501-41313035 | ||||||||||||||||||||||
| Strand | Reverse strand | ||||||||||||||||||||||
| Band | |||||||||||||||||||||||
| Transcripts |
|
||||||||||||||||||||||
| Interactions | |||||||||||||||||||||||
| Number of Interactions |
This gene and/or its encoded proteins are associated with 0 experimentally validated interaction(s) in this database.
They are also associated with 2 interaction(s) predicted by orthology.
|
||||||||||||||||||||||
| Gene Ontology | |||||||||||||||||||||||
Molecular Function |
|
||||||||||||||||||||||
| Biological Process |
|
||||||||||||||||||||||
| Cellular Component |
|
||||||||||||||||||||||
| Orthologs | |||||||||||||||||||||||
|
Species
Homo sapiens
Mus musculus
|
Gene ID
Gene Order
|
||||||||||||||||||||||
| Pathways | |||||||||||||||||||||||
| NETPATH | |||||||||||||||||||||||
| REACTOME |
Signaling by NOTCH pathway
Pre-NOTCH Expression and Processing pathway
Signal Transduction pathway
Pre-NOTCH Processing in Golgi pathway
|
||||||||||||||||||||||
| KEGG | |||||||||||||||||||||||
| INOH | |||||||||||||||||||||||
| PID NCI | |||||||||||||||||||||||
| Pathway Predictions based on Human Orthology Data | |||||||||||||||||||||||
| NETPATH |
Notch pathway
|
||||||||||||||||||||||
| REACTOME |
Pre-NOTCH Processing in Golgi pathway
Pre-NOTCH Expression and Processing pathway
Signal Transduction pathway
Signaling by NOTCH pathway
Pre-NOTCH Expression and Processing pathway
Pre-NOTCH Processing in Golgi pathway
Signal Transduction pathway
Signaling by NOTCH pathway
|
||||||||||||||||||||||
| KEGG |
Notch signaling pathway pathway
Other types of O-glycan biosynthesis pathway
Notch signaling pathway pathway
Other types of O-glycan biosynthesis pathway
|
||||||||||||||||||||||
| INOH | |||||||||||||||||||||||
| PID NCI | |||||||||||||||||||||||
| Cross-References | |||||||||||||||||||||||
| SwissProt | |||||||||||||||||||||||
| TrEMBL | |||||||||||||||||||||||
| UniProt Splice Variant | |||||||||||||||||||||||
| Entrez Gene | |||||||||||||||||||||||
| UniGene | Bt.11476 | ||||||||||||||||||||||
| RefSeq | NM_001046222 | ||||||||||||||||||||||
| HUGO | |||||||||||||||||||||||
| OMIM | |||||||||||||||||||||||
| CCDS | |||||||||||||||||||||||
| HPRD | |||||||||||||||||||||||
| IMGT | |||||||||||||||||||||||
| EMBL | |||||||||||||||||||||||
| GenPept | |||||||||||||||||||||||
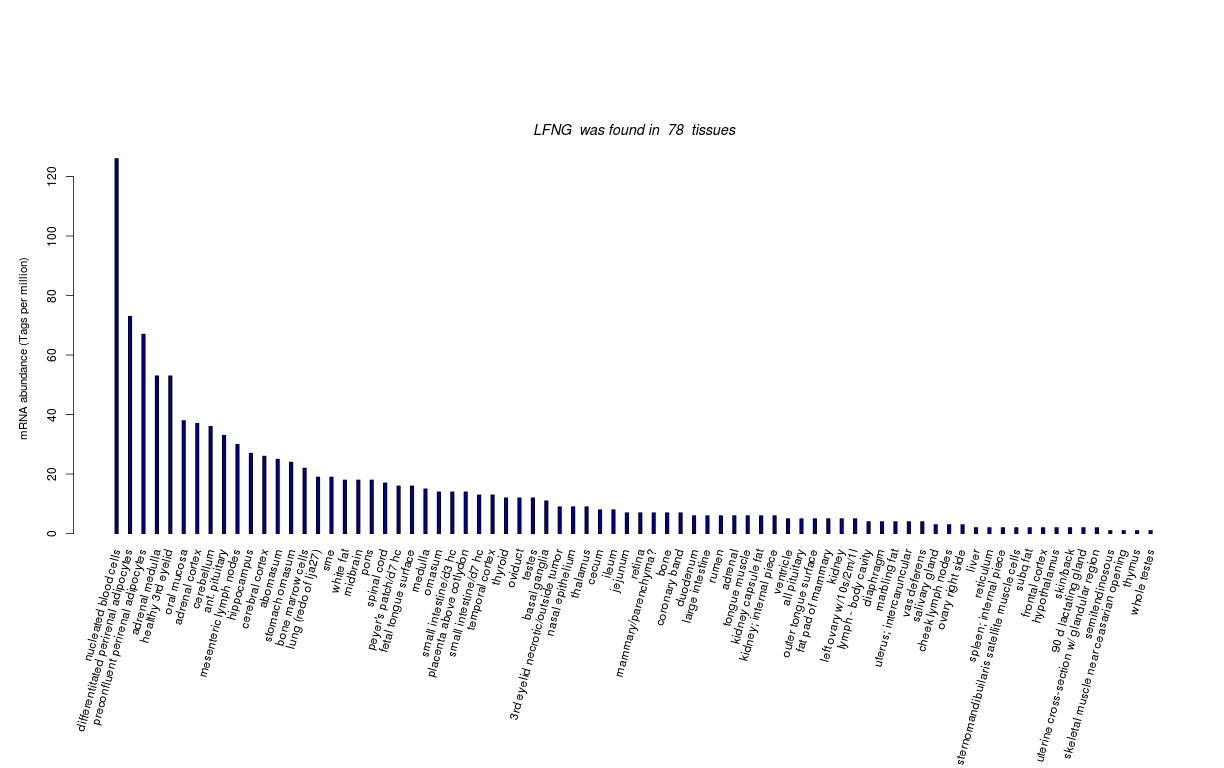
| RNA Seq Atlas | |||||||||||||||||||||||
| Transcript Frequencies | |||||||||||||||||||||||
| Tag Count based mRNA-Abundances across 87 different Tissues (TPM).
Based on Data from Bovine Gene Atlas |
(Move your mouse over the image to view a more detailed version) |
||||||||||||||||||||||