| Bos taurus Gene: FZD4 | |||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Summary | |||||||||||||||||||||||||||||||||||||||||||
| InnateDB Gene | IDBG-641882.3 | ||||||||||||||||||||||||||||||||||||||||||
| Last Modified | 2014-10-13 [Report errors or provide feedback] | ||||||||||||||||||||||||||||||||||||||||||
| Gene Symbol | FZD4 | ||||||||||||||||||||||||||||||||||||||||||
| Gene Name | frizzled-4 precursor | ||||||||||||||||||||||||||||||||||||||||||
| Synonyms | |||||||||||||||||||||||||||||||||||||||||||
| Species | Bos taurus | ||||||||||||||||||||||||||||||||||||||||||
| Ensembl Gene | ENSBTAG00000040128 | ||||||||||||||||||||||||||||||||||||||||||
| Encoded Proteins |
Uncharacterized protein
|
||||||||||||||||||||||||||||||||||||||||||
| Protein Structure | |||||||||||||||||||||||||||||||||||||||||||
| Useful resources | Stemformatics EHFPI ImmGen | ||||||||||||||||||||||||||||||||||||||||||
| Entrez Gene | |||||||||||||||||||||||||||||||||||||||||||
| Summary |
This gene does not have any Entrez summary - the following is the summary from its human ortholog ENSG00000174804:
This gene is a member of the frizzled gene family. Members of this family encode seven-transmembrane domain proteins that are receptors for the Wingless type MMTV integration site family of signaling proteins. Most frizzled receptors are coupled to the beta-catenin canonical signaling pathway. This protein may play a role as a positive regulator of the Wingless type MMTV integration site signaling pathway. A transcript variant retaining intronic sequence and encoding a shorter isoform has been described, however, its expression is not supported by other experimental evidence. [provided by RefSeq, Jul 2008] |
||||||||||||||||||||||||||||||||||||||||||
| Gene Information | |||||||||||||||||||||||||||||||||||||||||||
| Type | Protein coding | ||||||||||||||||||||||||||||||||||||||||||
| Genomic Location | Chromosome 29:8669606-8673380 | ||||||||||||||||||||||||||||||||||||||||||
| Strand | Forward strand | ||||||||||||||||||||||||||||||||||||||||||
| Band | |||||||||||||||||||||||||||||||||||||||||||
| Transcripts |
|
||||||||||||||||||||||||||||||||||||||||||
| Interactions | |||||||||||||||||||||||||||||||||||||||||||
| Number of Interactions |
This gene and/or its encoded proteins are associated with 0 experimentally validated interaction(s) in this database.
They are also associated with 13 interaction(s) predicted by orthology.
|
||||||||||||||||||||||||||||||||||||||||||
| Gene Ontology | |||||||||||||||||||||||||||||||||||||||||||
Molecular Function |
|
||||||||||||||||||||||||||||||||||||||||||
| Biological Process |
|
||||||||||||||||||||||||||||||||||||||||||
| Cellular Component |
|
||||||||||||||||||||||||||||||||||||||||||
| Orthologs | |||||||||||||||||||||||||||||||||||||||||||
|
Species
Homo sapiens
Mus musculus
|
Gene ID
Gene Order
|
||||||||||||||||||||||||||||||||||||||||||
| Pathway Predictions based on Human Orthology Data | |||||||||||||||||||||||||||||||||||||||||||
| NETPATH |
Wnt pathway
|
||||||||||||||||||||||||||||||||||||||||||
| REACTOME |
Class B/2 (Secretin family receptors) pathway
PCP/CE pathway pathway
Ca2+ pathway pathway
misspliced LRP5 mutants have enhanced beta-catenin-dependent signaling pathway
WNT5A-dependent internalization of FZD4 pathway
Signaling by WNT in cancer pathway
beta-catenin independent WNT signaling pathway
Signaling by Wnt pathway
Signaling by GPCR pathway
Signal Transduction pathway
regulation of FZD by ubiquitination pathway
TCF dependent signaling in response to WNT pathway
Asymmetric localization of PCP proteins pathway
RNF mutants show enhanced WNT signaling and proliferation pathway
GPCR ligand binding pathway
XAV939 inhibits tankyrase, stabilizing AXIN pathway
Disease pathway
WNT5A-dependent internalization of FZD4 pathway
Disease pathway
Signaling by Wnt pathway
Signaling by GPCR pathway
Class B/2 (Secretin family receptors) pathway
Signaling by WNT in cancer pathway
beta-catenin independent WNT signaling pathway
regulation of FZD by ubiquitination pathway
misspliced LRP5 mutants have enhanced beta-catenin-dependent signaling pathway
PCP/CE pathway pathway
RNF mutants show enhanced WNT signaling and proliferation pathway
TCF dependent signaling in response to WNT pathway
GPCR ligand binding pathway
Ca2+ pathway pathway
Signal Transduction pathway
XAV939 inhibits tankyrase, stabilizing AXIN pathway
|
||||||||||||||||||||||||||||||||||||||||||
| KEGG |
Wnt signaling pathway pathway
Basal cell carcinoma pathway
Melanogenesis pathway
Pathways in cancer pathway
Basal cell carcinoma pathway
Melanogenesis pathway
Wnt signaling pathway pathway
Pathways in cancer pathway
|
||||||||||||||||||||||||||||||||||||||||||
| INOH |
Wnt signaling pathway pathway
GPCR signaling pathway
Wnt signaling pathway pathway
GPCR signaling pathway
|
||||||||||||||||||||||||||||||||||||||||||
| PID NCI |
Wnt signaling network
|
||||||||||||||||||||||||||||||||||||||||||
| Cross-References | |||||||||||||||||||||||||||||||||||||||||||
| SwissProt | |||||||||||||||||||||||||||||||||||||||||||
| TrEMBL | F1MLJ8 Q1MW19 | ||||||||||||||||||||||||||||||||||||||||||
| UniProt Splice Variant | |||||||||||||||||||||||||||||||||||||||||||
| Entrez Gene | 445416 | ||||||||||||||||||||||||||||||||||||||||||
| UniGene | Bt.76547 | ||||||||||||||||||||||||||||||||||||||||||
| RefSeq | NM_001206269 | ||||||||||||||||||||||||||||||||||||||||||
| HUGO | |||||||||||||||||||||||||||||||||||||||||||
| OMIM | |||||||||||||||||||||||||||||||||||||||||||
| CCDS | |||||||||||||||||||||||||||||||||||||||||||
| HPRD | |||||||||||||||||||||||||||||||||||||||||||
| IMGT | |||||||||||||||||||||||||||||||||||||||||||
| EMBL | AB257753 DAAA02062527 | ||||||||||||||||||||||||||||||||||||||||||
| GenPept | BAE93504 | ||||||||||||||||||||||||||||||||||||||||||
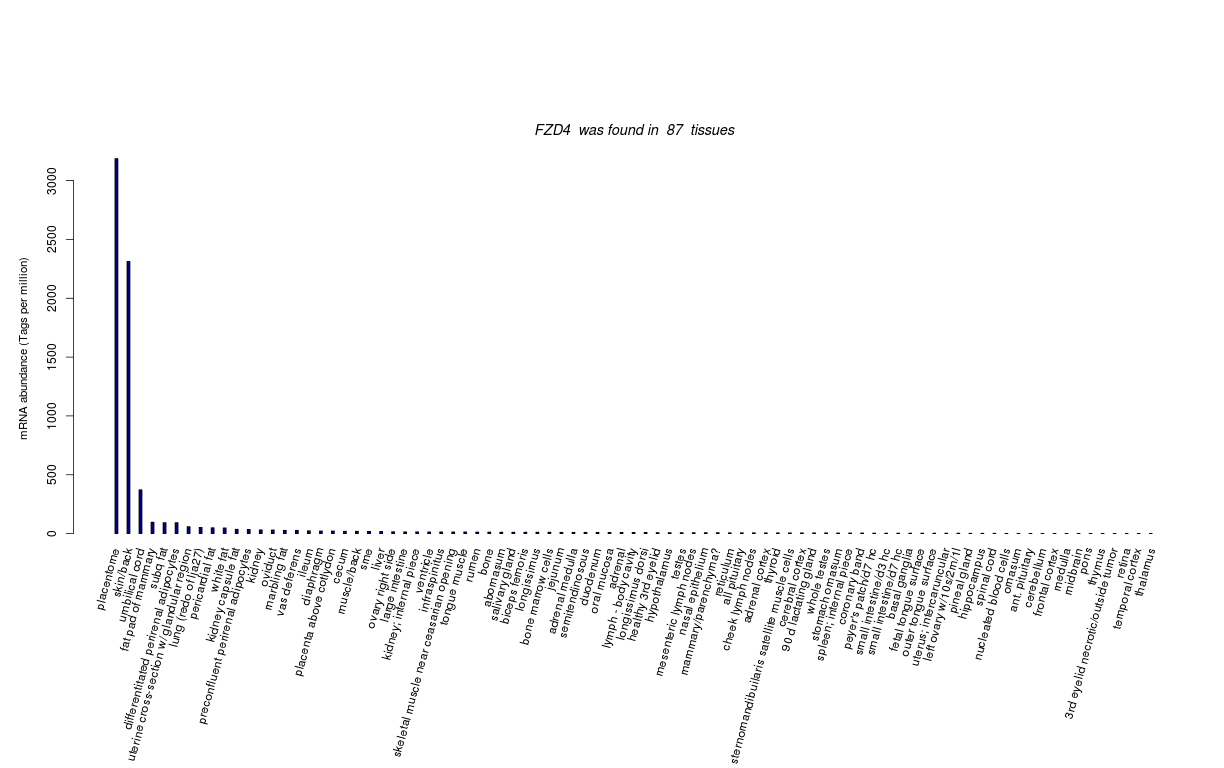
| RNA Seq Atlas | 445416 | ||||||||||||||||||||||||||||||||||||||||||
| Transcript Frequencies | |||||||||||||||||||||||||||||||||||||||||||
| Tag Count based mRNA-Abundances across 87 different Tissues (TPM).
Based on Data from Bovine Gene Atlas |
(Move your mouse over the image to view a more detailed version) |
||||||||||||||||||||||||||||||||||||||||||