| Bos taurus Gene: ILF3 | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Summary | |||||||||||||
| InnateDB Gene | IDBG-643329.3 | ||||||||||||
| Last Modified | 2014-10-13 [Report errors or provide feedback] | ||||||||||||
| Gene Symbol | ILF3 | ||||||||||||
| Gene Name | interleukin enhancer-binding factor 3 | ||||||||||||
| Synonyms | |||||||||||||
| Species | Bos taurus | ||||||||||||
| Ensembl Gene | ENSBTAG00000040076 | ||||||||||||
| Encoded Proteins |
interleukin enhancer binding factor 3, 90kDa
|
||||||||||||
| Protein Structure | |||||||||||||
| Useful resources | Stemformatics EHFPI ImmGen | ||||||||||||
| InnateDB Annotation from Orthologs | |||||||||||||
| Summary |
[Homo sapiens] ILF3 is an RNA-binding protein that influences mRNA turnover and/or translation by regulating mRNA stability. ILF3 can bind to mitogen-activated protein (MAP) kinase phosphatase 1 (MKP-1) and increase its mRNA stability and translation, resulting in increased MKP-1 dephosphorylation activity and thereby inactivation of MAP kinases extracellular signal-regulated kinase (ERK), c-Jun N-terminal kinase (JNK), and p38.
|
||||||||||||
| Entrez Gene | |||||||||||||
| Summary |
This gene does not have any Entrez summary - the following is the summary from its human ortholog ENSG00000129351:
This gene encodes a double-stranded RNA (dsRNA) binding protein that complexes with other proteins, dsRNAs, small noncoding RNAs, and mRNAs to regulate gene expression and stabilize mRNAs. This protein was first discovered to be a subunit of the nuclear factor of activated T-cells (NFAT); a transcription factor required for T-cell expression of interleukin 2. NFAT is a heterodimer of 45 kDa and 90 kDa proteins, the larger of which is the product of this gene. These proteins have been shown to affect the redistribution of nuclear mRNA to the cytoplasm. Knockdown of NF45 or NF90 protein retards cell growth; possibly by inhibition of mRNA stabilization. In contrast, an isoform (NF110) of this gene that is predominantly restricted to the nucleus has only minor effects on cell growth when its levels are reduced. Alternative splicing results in multiple transcript variants encoding distinct isoforms.[provided by RefSeq, Nov 2008] |
||||||||||||
| Gene Information | |||||||||||||
| Type | Protein coding | ||||||||||||
| Genomic Location | Chromosome 7:16375687-16391318 | ||||||||||||
| Strand | Forward strand | ||||||||||||
| Band | |||||||||||||
| Transcripts |
|
||||||||||||
| Interactions | |||||||||||||
| Number of Interactions |
This gene and/or its encoded proteins are associated with 0 experimentally validated interaction(s) in this database.
They are also associated with 231 interaction(s) predicted by orthology.
|
||||||||||||
| Gene Ontology | |||||||||||||
Molecular Function |
|
||||||||||||
| Biological Process |
|
||||||||||||
| Cellular Component |
|
||||||||||||
| Orthologs | |||||||||||||
|
Species
Homo sapiens
Mus musculus
|
Gene ID
Gene Order
|
||||||||||||
| Cross-References | |||||||||||||
| SwissProt | |||||||||||||
| TrEMBL | |||||||||||||
| UniProt Splice Variant | |||||||||||||
| Entrez Gene | |||||||||||||
| UniGene | Bt.106779 | ||||||||||||
| RefSeq | NM_001206495 XM_005208768 | ||||||||||||
| HUGO | |||||||||||||
| OMIM | |||||||||||||
| CCDS | |||||||||||||
| HPRD | |||||||||||||
| IMGT | |||||||||||||
| EMBL | |||||||||||||
| GenPept | |||||||||||||
| RNA Seq Atlas | |||||||||||||
| Transcript Frequencies | |||||||||||||
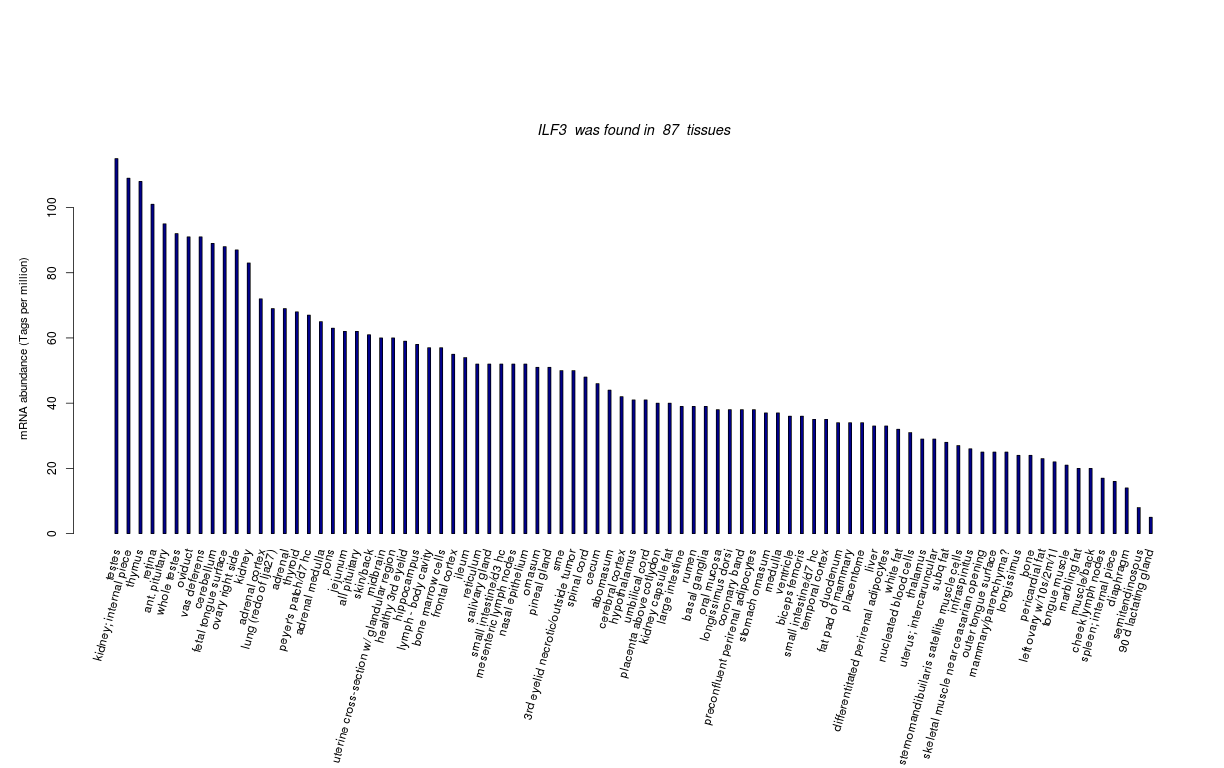
| Tag Count based mRNA-Abundances across 87 different Tissues (TPM).
Based on Data from Bovine Gene Atlas |
(Move your mouse over the image to view a more detailed version) |
||||||||||||