| Bos taurus Gene: BT.37181 | |||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Summary | |||||||||||||||||||||||||||||||||||||||
| InnateDB Gene | IDBG-646644.3 | ||||||||||||||||||||||||||||||||||||||
| Last Modified | 2014-10-13 [Report errors or provide feedback] | ||||||||||||||||||||||||||||||||||||||
| Gene Symbol | BT.37181 | ||||||||||||||||||||||||||||||||||||||
| Gene Name | hepatocyte nuclear factor 6 | ||||||||||||||||||||||||||||||||||||||
| Synonyms | HNF-6a | ||||||||||||||||||||||||||||||||||||||
| Species | Bos taurus | ||||||||||||||||||||||||||||||||||||||
| Ensembl Gene | ENSBTAG00000046470 | ||||||||||||||||||||||||||||||||||||||
| Encoded Proteins |
Uncharacterized protein
|
||||||||||||||||||||||||||||||||||||||
| Protein Structure | |||||||||||||||||||||||||||||||||||||||
| Useful resources | Stemformatics EHFPI ImmGen | ||||||||||||||||||||||||||||||||||||||
| Entrez Gene | |||||||||||||||||||||||||||||||||||||||
| Summary |
This gene does not have any Entrez summary - the following is the summary from its human ortholog ENSG00000169856:
This gene encodes a member of the Cut homeobox family of transcription factors. Expression of the encoded protein is enriched in the liver, where it stimulates transcription of liver-expressed genes, and antagonizes glucocorticoid-stimulated gene transcription. This gene may influence a variety of cellular processes including glucose metabolism, cell cycle regulation, and it may also be associated with cancer. Alternative splicing results in multiple transcript variants. [provided by RefSeq, Dec 2012] |
||||||||||||||||||||||||||||||||||||||
| Gene Information | |||||||||||||||||||||||||||||||||||||||
| Type | Protein coding | ||||||||||||||||||||||||||||||||||||||
| Genomic Location | Chromosome 10:57712108-57744837 | ||||||||||||||||||||||||||||||||||||||
| Strand | Forward strand | ||||||||||||||||||||||||||||||||||||||
| Band | |||||||||||||||||||||||||||||||||||||||
| Transcripts |
|
||||||||||||||||||||||||||||||||||||||
| Interactions | |||||||||||||||||||||||||||||||||||||||
| Number of Interactions |
This gene and/or its encoded proteins are associated with 0 experimentally validated interaction(s) in this database.
They are also associated with 210 interaction(s) predicted by orthology.
|
||||||||||||||||||||||||||||||||||||||
| Gene Ontology | |||||||||||||||||||||||||||||||||||||||
Molecular Function |
|
||||||||||||||||||||||||||||||||||||||
| Biological Process |
|
||||||||||||||||||||||||||||||||||||||
| Cellular Component |
|
||||||||||||||||||||||||||||||||||||||
| Orthologs | |||||||||||||||||||||||||||||||||||||||
|
Species
Homo sapiens
Mus musculus
|
Gene ID
Gene Order
|
||||||||||||||||||||||||||||||||||||||
| Pathway Predictions based on Human Orthology Data | |||||||||||||||||||||||||||||||||||||||
| NETPATH | |||||||||||||||||||||||||||||||||||||||
| REACTOME |
Regulation of gene expression in late stage (branching morphogenesis) pancreatic bud precursor cells pathway
Regulation of gene expression in early pancreatic precursor cells pathway
Developmental Biology pathway
Regulation of beta-cell development pathway
Regulation of gene expression in early pancreatic precursor cells pathway
Regulation of gene expression in late stage (branching morphogenesis) pancreatic bud precursor cells pathway
Developmental Biology pathway
Regulation of beta-cell development pathway
|
||||||||||||||||||||||||||||||||||||||
| KEGG |
Maturity onset diabetes of the young pathway
Maturity onset diabetes of the young pathway
|
||||||||||||||||||||||||||||||||||||||
| INOH | |||||||||||||||||||||||||||||||||||||||
| PID NCI |
FOXM1 transcription factor network
|
||||||||||||||||||||||||||||||||||||||
| Cross-References | |||||||||||||||||||||||||||||||||||||||
| SwissProt | |||||||||||||||||||||||||||||||||||||||
| TrEMBL | G3N248 | ||||||||||||||||||||||||||||||||||||||
| UniProt Splice Variant | |||||||||||||||||||||||||||||||||||||||
| Entrez Gene | 503584 | ||||||||||||||||||||||||||||||||||||||
| UniGene | Bt.106196 Bt.37181 | ||||||||||||||||||||||||||||||||||||||
| RefSeq | NM_001191161 | ||||||||||||||||||||||||||||||||||||||
| HUGO | |||||||||||||||||||||||||||||||||||||||
| OMIM | |||||||||||||||||||||||||||||||||||||||
| CCDS | |||||||||||||||||||||||||||||||||||||||
| HPRD | |||||||||||||||||||||||||||||||||||||||
| IMGT | |||||||||||||||||||||||||||||||||||||||
| EMBL | DAAA02028984 | ||||||||||||||||||||||||||||||||||||||
| GenPept | |||||||||||||||||||||||||||||||||||||||
| RNA Seq Atlas | 503584 | ||||||||||||||||||||||||||||||||||||||
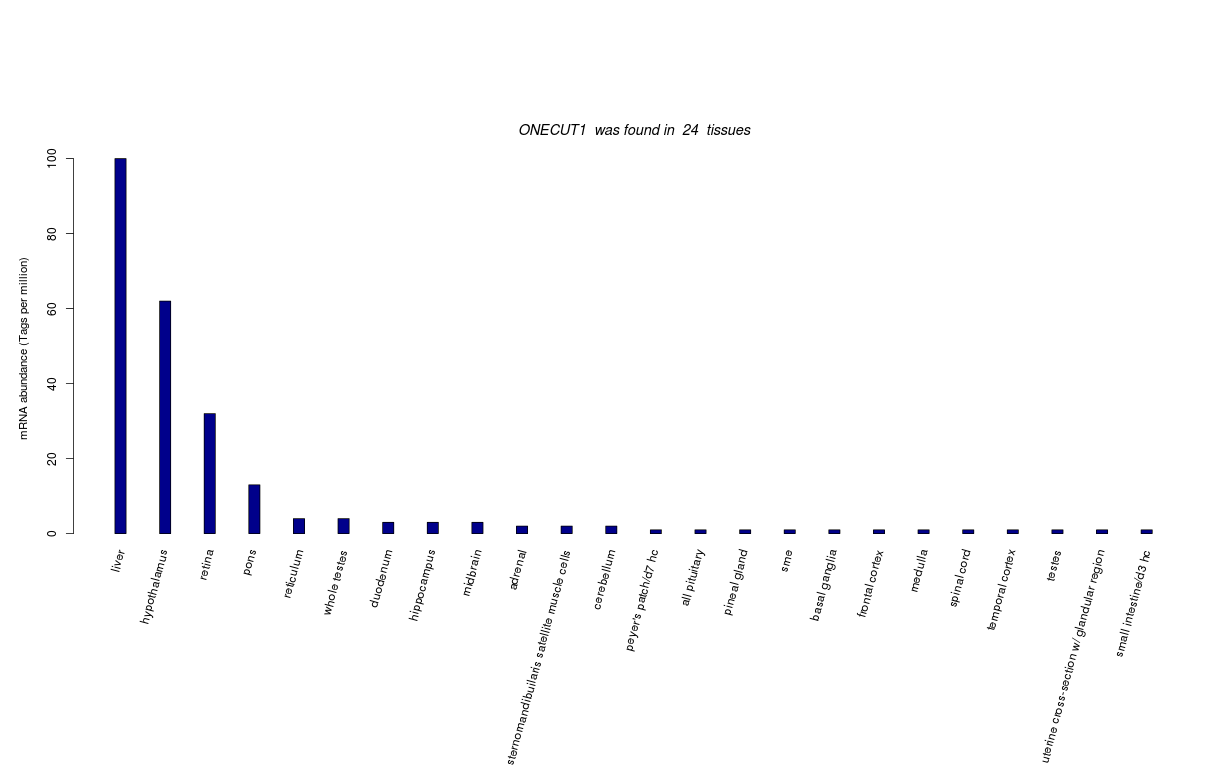
| Transcript Frequencies | |||||||||||||||||||||||||||||||||||||||
| Tag Count based mRNA-Abundances across 87 different Tissues (TPM).
Based on Data from Bovine Gene Atlas |
(Move your mouse over the image to view a more detailed version) |
||||||||||||||||||||||||||||||||||||||