| Bos taurus Gene: CITED1 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Summary | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| InnateDB Gene | IDBG-635694.3 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Last Modified | 2014-10-13 [Report errors or provide feedback] | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Gene Symbol | CITED1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Gene Name | Cbp/p300-interacting transactivator 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Synonyms | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Species | Bos taurus | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Ensembl Gene | ENSBTAG00000045925 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Encoded Proteins |
Cbp/p300-interacting transactivator 1
Cbp/p300-interacting transactivator 1
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Protein Structure |

|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Useful resources | Stemformatics EHFPI ImmGen | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Entrez Gene | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Summary |
This gene does not have any Entrez summary - the following is the summary from its human ortholog ENSG00000125931:
This gene encodes a member of the CREB-binding protein/p300-interacting transactivator with Asp/Glu-rich C-terminal domain (CITED) family of proteins. The encoded protein, also known as melanocyte-specific gene 1, may function as a transcriptional coactivator and may play a role in pigmentation of melanocytes. Alternatively spliced transcript variants have been described. [provided by RefSeq, Jan 2009] |
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Gene Information | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Type | Protein coding | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Genomic Location | Chromosome X:83495862-83501893 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Strand | Forward strand | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Band | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Transcripts |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Interactions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Number of Interactions |
This gene and/or its encoded proteins are associated with 0 experimentally validated interaction(s) in this database.
They are also associated with 16 interaction(s) predicted by orthology.
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Gene Ontology | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Molecular Function |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Biological Process |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Cellular Component |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Orthologs | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
Species
Homo sapiens
Mus musculus
|
Gene ID
Gene Order
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Pathway Predictions based on Human Orthology Data | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| NETPATH |
TGF_beta_Receptor pathway
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| REACTOME | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| KEGG | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| INOH | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| PID NCI |
Regulation of nuclear SMAD2/3 signaling
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Cross-References | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| SwissProt | Q9BDI3 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| TrEMBL | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| UniProt Splice Variant | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Entrez Gene | 282182 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| UniGene | Bt.4437 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| RefSeq | XM_005227988 XM_005227989 XM_005227991 NM_174518 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| HUGO | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| OMIM | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| CCDS | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| HPRD | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| IMGT | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| EMBL | AF362075 BC102725 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| GenPept | AAI02726 AAK27241 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| RNA Seq Atlas | 282182 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Transcript Frequencies | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
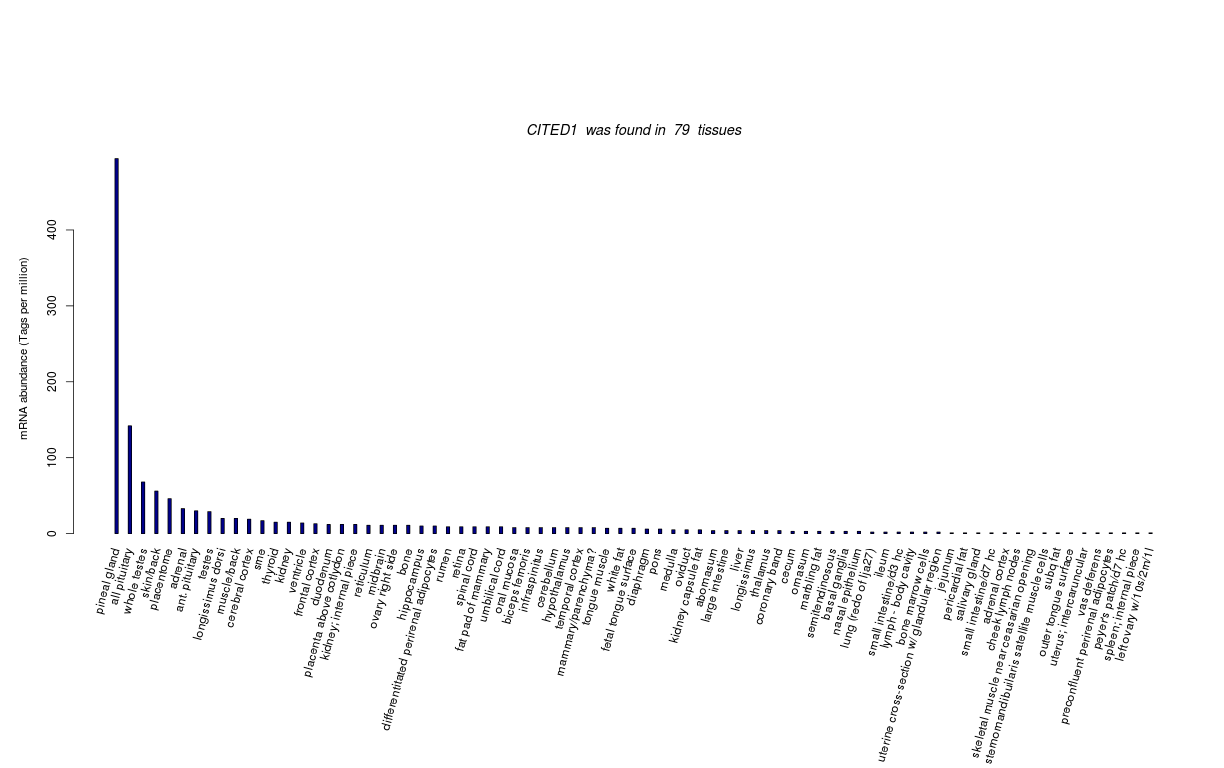
| Tag Count based mRNA-Abundances across 87 different Tissues (TPM).
Based on Data from Bovine Gene Atlas |
(Move your mouse over the image to view a more detailed version) |
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||