| Bos taurus Gene: NOTCH3 | |||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Summary | |||||||||||||||||||||
| InnateDB Gene | IDBG-642521.3 | ||||||||||||||||||||
| Last Modified | 2014-10-13 [Report errors or provide feedback] | ||||||||||||||||||||
| Gene Symbol | NOTCH3 | ||||||||||||||||||||
| Gene Name | Uncharacterized protein | ||||||||||||||||||||
| Synonyms | |||||||||||||||||||||
| Species | Bos taurus | ||||||||||||||||||||
| Ensembl Gene | ENSBTAG00000043971 | ||||||||||||||||||||
| Encoded Proteins |
notch 3
|
||||||||||||||||||||
| Protein Structure | |||||||||||||||||||||
| Useful resources | Stemformatics EHFPI ImmGen | ||||||||||||||||||||
| Entrez Gene | |||||||||||||||||||||
| Summary |
This gene does not have any Entrez summary - the following is the summary from its human ortholog ENSG00000074181:
This gene encodes the third discovered human homologue of the Drosophilia melanogaster type I membrane protein notch. In Drosophilia, notch interaction with its cell-bound ligands (delta, serrate) establishes an intercellular signalling pathway that plays a key role in neural development. Homologues of the notch-ligands have also been identified in human, but precise interactions between these ligands and the human notch homologues remains to be determined. Mutations in NOTCH3 have been identified as the underlying cause of cerebral autosomal dominant arteriopathy with subcortical infarcts and leukoencephalopathy (CADASIL). [provided by RefSeq, Jul 2008] |
||||||||||||||||||||
| Gene Information | |||||||||||||||||||||
| Type | Protein coding | ||||||||||||||||||||
| Genomic Location | Chromosome 7:8930803-8970307 | ||||||||||||||||||||
| Strand | Forward strand | ||||||||||||||||||||
| Band | |||||||||||||||||||||
| Transcripts |
|
||||||||||||||||||||
| Interactions | |||||||||||||||||||||
| Number of Interactions |
This gene and/or its encoded proteins are associated with 0 experimentally validated interaction(s) in this database.
They are also associated with 20 interaction(s) predicted by orthology.
|
||||||||||||||||||||
| Gene Ontology | |||||||||||||||||||||
Molecular Function |
|
||||||||||||||||||||
| Biological Process |
|
||||||||||||||||||||
| Cellular Component |
|
||||||||||||||||||||
| Orthologs | |||||||||||||||||||||
|
Species
Homo sapiens
Mus musculus
|
Gene ID
Gene Order
|
||||||||||||||||||||
| Pathway Predictions based on Human Orthology Data | |||||||||||||||||||||
| NETPATH |
Notch pathway
|
||||||||||||||||||||
| REACTOME |
Signaling by NOTCH3 pathway
Pre-NOTCH Transcription and Translation pathway
Pre-NOTCH Processing in the Endoplasmic Reticulum pathway
Pre-NOTCH Processing in Golgi pathway
Notch-HLH transcription pathway pathway
Generic Transcription Pathway pathway
Pre-NOTCH Expression and Processing pathway
Signal Transduction pathway
Signaling by NOTCH pathway
Gene Expression pathway
Notch-HLH transcription pathway pathway
Signaling by NOTCH3 pathway
Pre-NOTCH Processing in the Endoplasmic Reticulum pathway
Pre-NOTCH Expression and Processing pathway
Gene Expression pathway
Pre-NOTCH Processing in Golgi pathway
Generic Transcription Pathway pathway
Signal Transduction pathway
Signaling by NOTCH pathway
|
||||||||||||||||||||
| KEGG |
Notch signaling pathway pathway
Dorso-ventral axis formation pathway
Notch signaling pathway pathway
Dorso-ventral axis formation pathway
|
||||||||||||||||||||
| INOH |
Notch signaling pathway pathway
Notch signaling pathway pathway
|
||||||||||||||||||||
| PID NCI |
Notch signaling pathway
|
||||||||||||||||||||
| Cross-References | |||||||||||||||||||||
| SwissProt | |||||||||||||||||||||
| TrEMBL | |||||||||||||||||||||
| UniProt Splice Variant | |||||||||||||||||||||
| Entrez Gene | |||||||||||||||||||||
| UniGene | Bt.111418 Bt.111718 Bt.111734 | ||||||||||||||||||||
| RefSeq | XM_003582382 XM_003586246 | ||||||||||||||||||||
| HUGO | |||||||||||||||||||||
| OMIM | |||||||||||||||||||||
| CCDS | |||||||||||||||||||||
| HPRD | |||||||||||||||||||||
| IMGT | |||||||||||||||||||||
| EMBL | |||||||||||||||||||||
| GenPept | |||||||||||||||||||||
| RNA Seq Atlas | |||||||||||||||||||||
| Transcript Frequencies | |||||||||||||||||||||
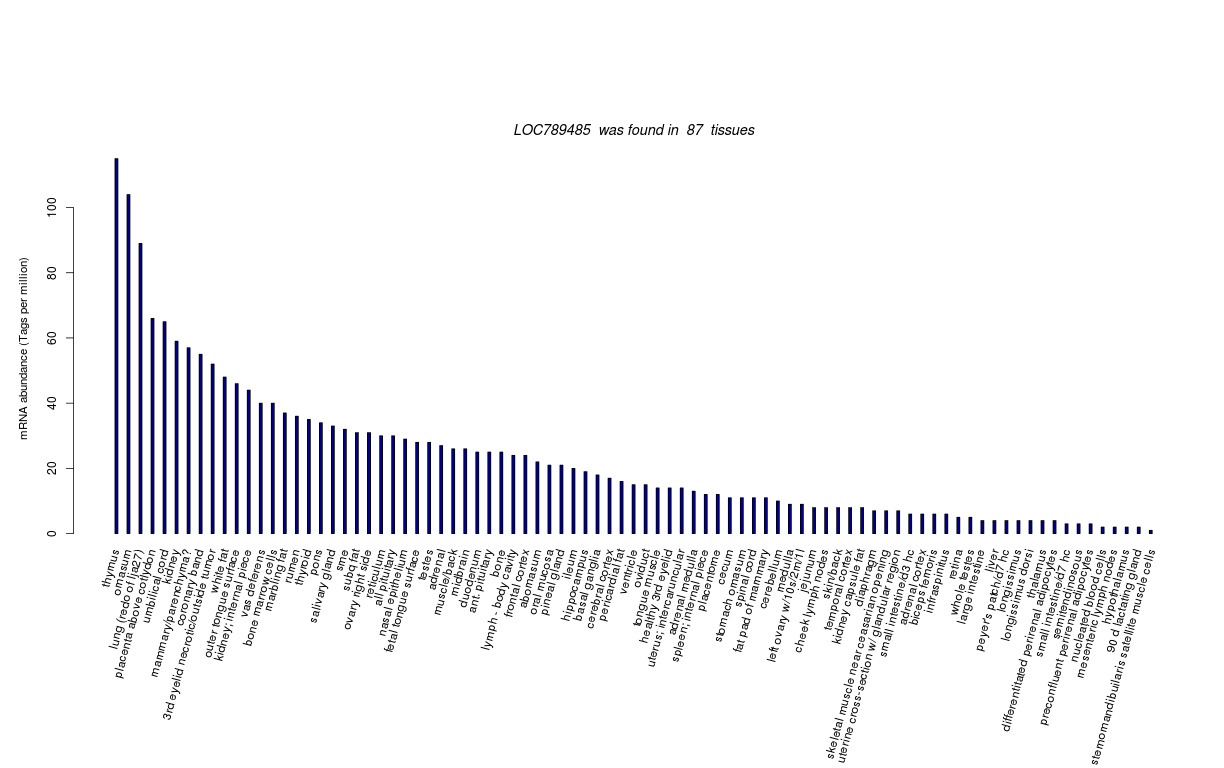
| Tag Count based mRNA-Abundances across 87 different Tissues (TPM).
Based on Data from Bovine Gene Atlas |
(Move your mouse over the image to view a more detailed version) |
||||||||||||||||||||